
 | Recent Developments in Amber Biomolecular Simulations Journal of Chemical Information and Modeling (2025) 65, 7835-7843 DOI: 10.1021/acs.jcim.5c01063 Amber is a molecular dynamics (MD) software package first conceived by Peter Kollman, his lab and collaborators to simulate biomolecular systems. The pmemd module is available as a serial version for central processing units (CPUs), NVIDIA and Advanced Micro Devices (AMD) graphics processing unit (GPU) versions as well as Message Passing Interface (MPI) parallel versions. Advanced capabilities include thermodynamic integration, replica exchange MD and accelerated MD methods. A brief update to the software and recently added capabilities is described in this Application Note. Read More View Full Article |
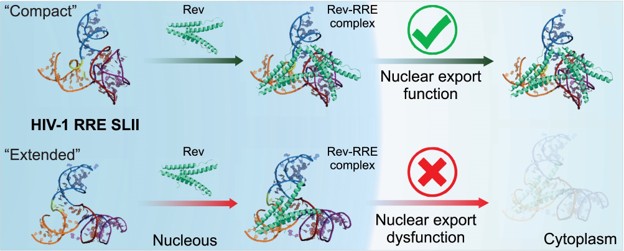 | Mutation-driven RRE stem-loop II conformational change induces HIV-1 nuclear export dysfunction Nucleic Acids Research (2025) 53, gkaf583 DOI: 10.1093/nar/gkaf583 The Rev response element (RRE) forms an oligomeric complex with the viral protein Rev to facilitate the nuclear export of intron-retaining viral RNAs during the late phase of HIV-1 (human immunodeficiency virus type 1) infection. However, the structures and mechanisms underlying this process remain largely unknown. Here, we determined the crystal structure of the HIV-1 RRE stem-loop II (SLII), revealing a unique three-way junction architecture in which the base stem (IIa) bifurcates into the stem-loops (IIb and IIc) to compose Rev binding sites. The crystal structures of various SLII mutants demonstrated that while some mutants retain the same “compact” fold as the wild type, other single-nucleotide mutants induce drastic conformational changes, forming an “extended” SLII structure. Through in vitro Rev binding assays and Rev activity measurements in HIV-1-infected cells using structure-guided SLII mutants designed to favor specific conformers, we showed that while the compact fold represents a functional SLII, the alternative extended conformation inhibits Rev binding and oligomerization and consequently stimulates HIV-1 RNA nuclear export dysfunction. The propensity of SLII to adopt multiple conformations as captured in crystal structures and their influence on Rev oligomerization illuminate emerging perspectives on RRE structural plasticity-based regulation of HIV-1 nuclear export and provide opportunities for developing anti-HIV drugs targeting specific RRE conformations. Read More View Full Article Download PDF |
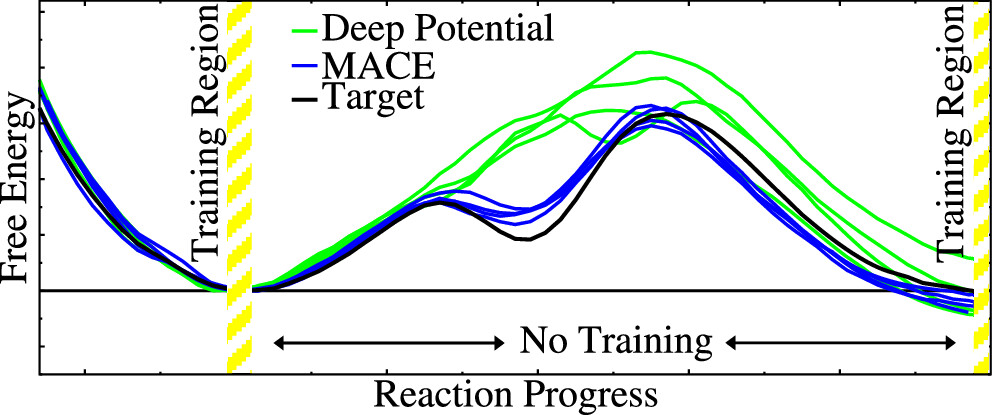 | Transferability of MACE Graph Neural Network for Range Corrected Δ-Machine Learning Potential QM/MM Applications The Journal of Physical Chemistry B (2025) 129, 5477-5490 DOI: 10.1021/acs.jpcb.5c02006 We previously introduced a “range corrected” Δ−machine learning potential (ΔMLP) that used deep neural networks to improve the accuracy of combined quantum mechanical/molecular mechanical (QM/MM) simulations by correcting both the internal QM and QM/MM interaction energies and forces [J. Chem. Theory Comput. 2021, 17, 6993–7009]. The present work extends this approach to include graph neural networks. Specifically, the approach is applied to the MACE message passing neural network architecture, and a series of AM1/d + MACE models are trained to reproduce PBE0/6–31G* QM/MM energies and forces of model phosphoryl transesterification reactions. Several models are designed to test the transferability of AM1/d + MACE by varying the amount of training data and calculating free energy surfaces of reactions that were not included in the parameter refinement. The transferability is compared to AM1/d + DP models that use the DeepPot-SE (DP) deep neural network architecture. The AM1/d + MACE models are found to reproduce the target free energy surfaces even in instances where the AM1/d + DP models exhibit inaccuracies. We train “end-state” models that include data only from the reactant and product states of the 6 reactions. Unlike the uncorrected AM1/d profiles, the AM1/d + MACE method correctly reproduces a stable pentacoordinated phosphorus intermediate even though the training did not include structures with a similar bonding pattern. Furthermore, the message passing mechanism hyperparameters defining the MACE network are varied to explore their effect on the model’s accuracy and performance. The AM1/d + MACE simulations are 28% slower than AM1/d QM/MM when the ΔMLP correction is performed on a graphics processing unit. Our results suggest that the MACE architecture may lead to ΔMLP models with improved transferability. Read More View Full Article Download PDF |
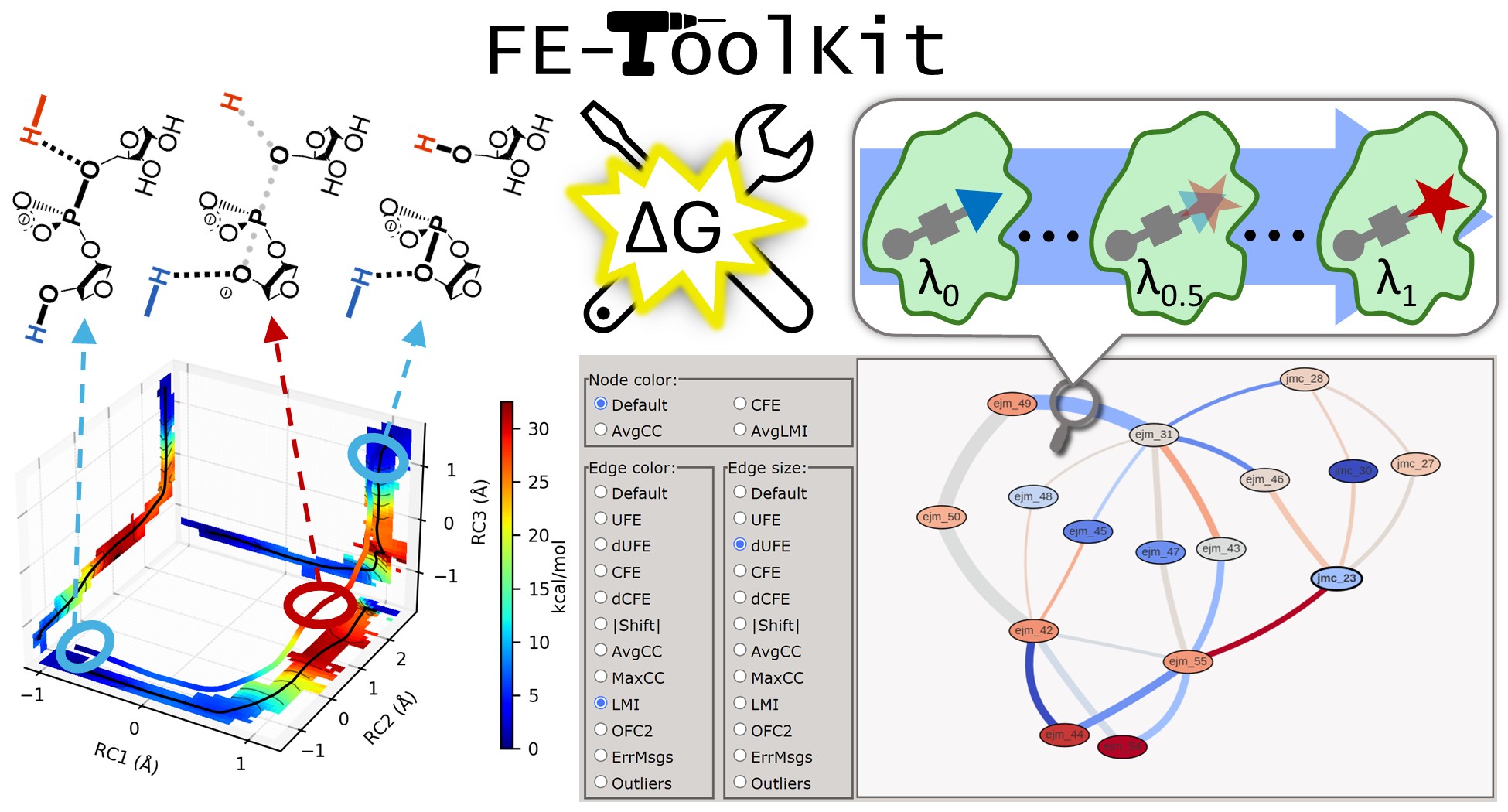 | FE-ToolKit: A Versatile Software Suite for Analysis of High-Dimensional Free Energy Surfaces and Alchemical Free Energy Networks Journal of Chemical Information and Modeling (2025) 65, 5273-5279 DOI: 10.1021/acs.jcim.5c00554 Free energy simulations play a pivotal role in diverse biological applications, including enzyme design, drug discovery, and biomolecular engineering. The characterization of high-dimensional free energy surfaces underlying complex enzymatic mechanisms necessitates extensive sampling through umbrella sampling or string method simulations. Accurate ranking of target-binding free energies across large ligand libraries relies on comprehensive alchemical free energy calculations organized into thermodynamic networks. The predictive accuracy of these methods hinges on robust, scalable tools for networkwide data analysis and extraction of physical properties from heterogeneous simulation data. Here, we introduce FE-ToolKit, a versatile software suite for the automated analysis of free energy surfaces, minimum free energy paths, and alchemical free energy networks (thermodynamic graphs). Read More View Full Article Download PDF |
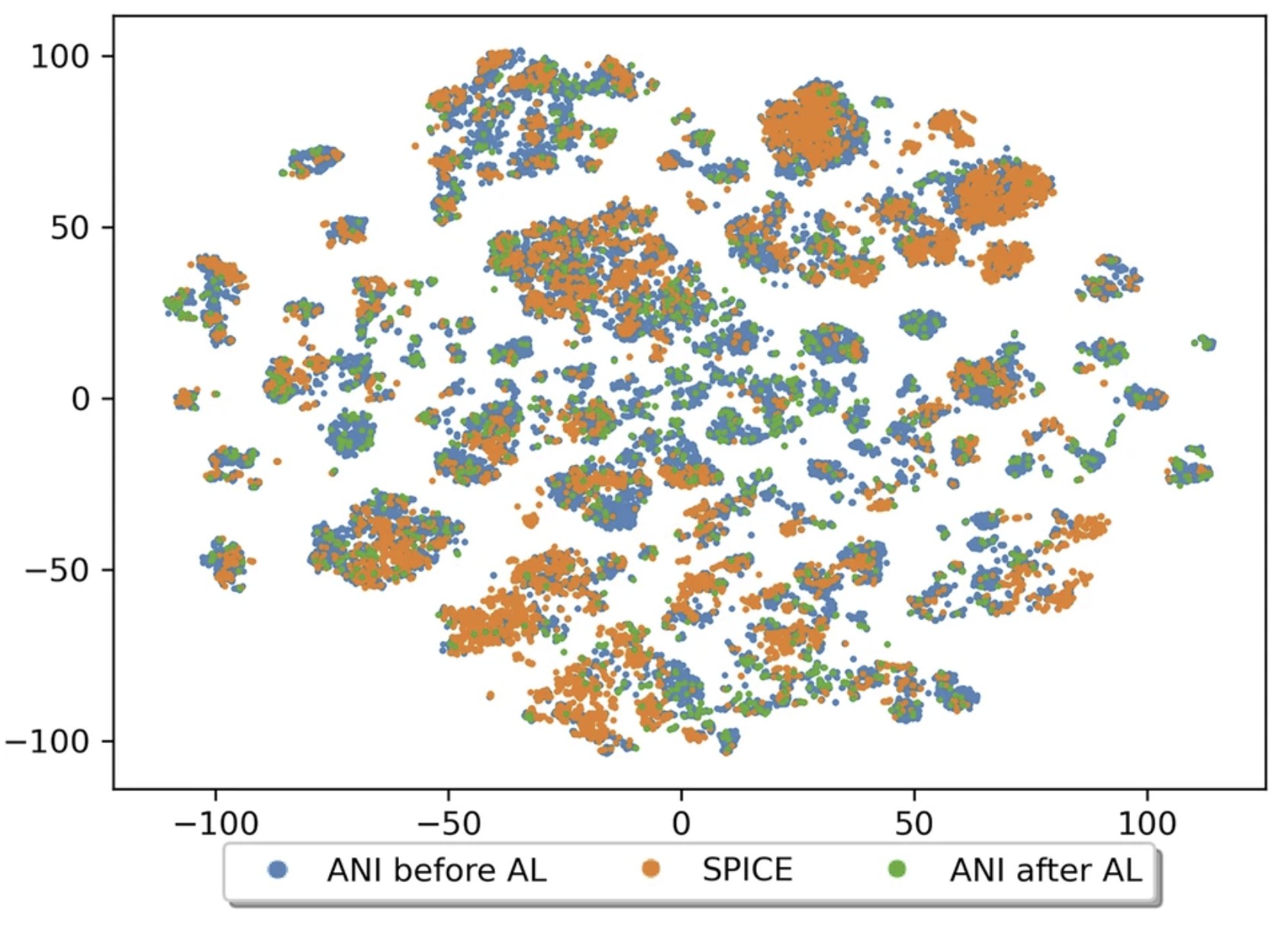 | The QDπ dataset, training data for drug-like molecules and biopolymer fragments and their interactions Scientific Data (2025) 12, 693 DOI: 10.1038/s41597-025-04972-3 The development of universal machine learning potentials (MLP) for small organic and drug-like molecules requires large, accurate datasets that span diverse chemical spaces. In this study, we introduce the QDπ dataset which incorporates data taken from several datasets. We use a query—by—committee active learning strategy to extract data from large datasets to maximize the diversity and avoid redundancy as relevant for neural network training to construct the QDπ dataset. The QDπ dataset requires only 1.6 million structures to express the chemical diversity of 13 elements from the various source datasets at the ωB97M-D3(BJ)/def2-TZVPPD level of theory. The QDπ dataset enables creation of flexible target loss functions for neural network training relevant to drug discovery, including information-dense data sets of relative conformational energies and barriers, intermolecular interactions, tautomers and relative protonation energies of drug-like compounds and biomolecular fragments. It is the hope that the high chemical information density and diversity contained in the QDπ dataset will provide a valuable resource for the development of new universal MLPs for drug discovery. Read More View Full Article Download PDF |
 | DeePMD-GNN: A DeePMD-kit Plugin for External Graph Neural Network Potentials Journal of Chemical Information and Modeling (2025) 65, 3154-3160 DOI: 10.1021/acs.jcim.4c02441 Machine learning potentials (MLPs) have revolutionized molecular simulation by providing efficient and accurate models for predicting atomic interactions. MLPs continue to advance and have had profound impact in applications that include drug discovery, enzyme catalysis, and materials design. The current landscape of MLP software presents challenges due to the limited interoperability between packages, which can lead to inconsistent benchmarking practices and necessitates separate interfaces with molecular dynamics (MD) software. To address these issues, we present DeePMD-GNN, a plugin for the DeePMD-kit framework that extends its capabilities to support external graph neural network (GNN) potentials.DeePMD-GNN enables the seamless integration of popular GNN-based models, such as NequIP and MACE, within the DeePMD-kit ecosystem. Furthermore, the new software infrastructure allows GNN models to be used within combined quantum mechanical/molecular mechanical (QM/MM) applications using the range corrected ΔMLP formalism.We demonstrate the application of DeePMD-GNN by performing benchmark calculations of NequIP, MACE, and DPA-2 models developed under consistent training conditions to ensure fair comparison. Read More View Full Article Download PDF |
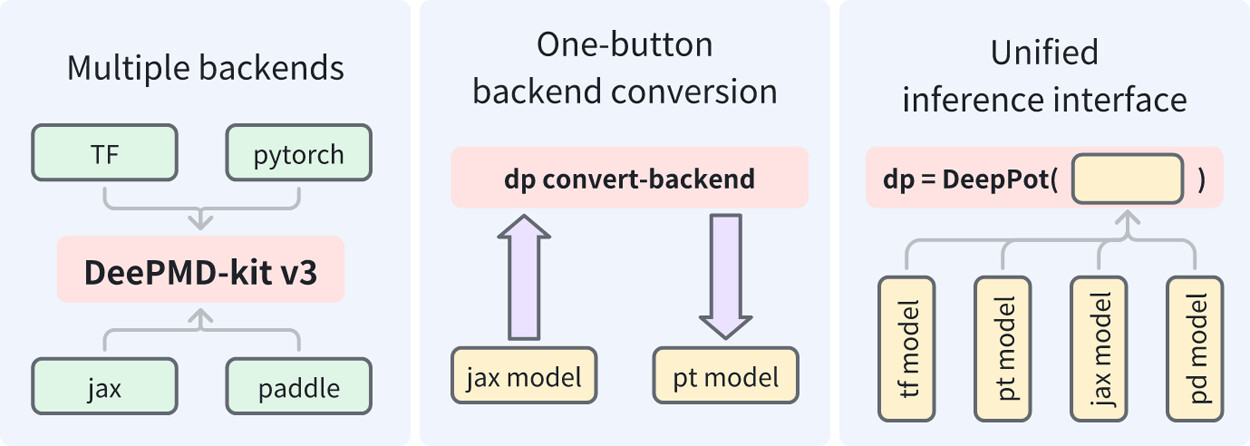 | DeePMD-kit v3: A Multiple-Backend Framework for Machine Learning Potentials Journal of Chemical Theory and Computation (2025) 21, 4375-4385 DOI: 10.1021/acs.jctc.5c00340 In recent years, machine learning potentials (MLPs) have become indispensable tools in physics, chemistry, and materials science, driving the development of software packages for molecular dynamics (MD) simulations and related applications. These packages, typically built on specific machine learning frameworks, such as TensorFlow, PyTorch, or JAX, face integration challenges when advanced applications demand communication across different frameworks. The previous TensorFlow-based implementation of the DeePMD-kit exemplified these limitations. In this work, we introduce DeePMD-kit version 3, a significant update featuring a multibackend framework that supports TensorFlow, PyTorch, JAX, and PaddlePaddle backends, and demonstrate the versatility of this architecture through the integration of other MLP packages and of differentiable molecular force fields. This architecture allows seamless back-end switching with minimal modifications, enabling users and developers to integrate DeePMD-kit with other packages using different machine learning frameworks. This innovation facilitates the development of more complex and interoperable workflows, paving the way for broader applications of MLPs in scientific research. Read More View Full Article |
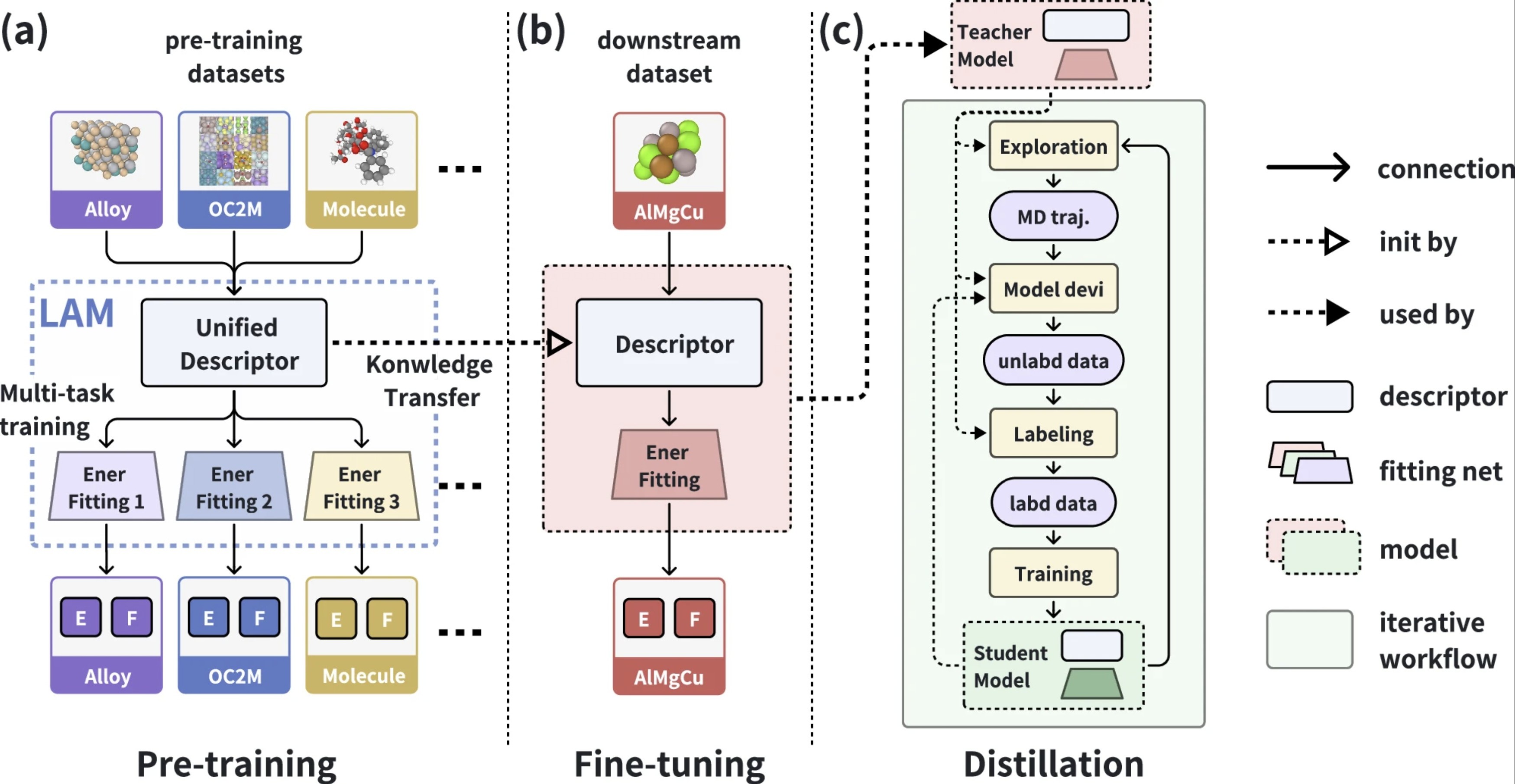 | DPA-2: a large atomic model as a multi-task learner npj Computational Materials (2024) 10, 293 DOI: 10.1038/s41524-024-01493-2 The rapid advancements in artificial intelligence (AI) are catalyzing transformative changes in atomic modeling, simulation, and design. AI-driven potential energy models have demonstrated the capability to conduct large-scale, long-duration simulations with the accuracy of ab initio electronic structure methods. However, the model generation process remains a bottleneck for large-scale applications. We propose a shift towards a model-centric ecosystem, wherein a large atomic model (LAM), pre-trained across multiple disciplines, can be efficiently fine-tuned and distilled for various downstream tasks, thereby establishing a new framework for molecular modeling. In this study, we introduce the DPA-2 architecture as a prototype for LAMs. Pre-trained on a diverse array of chemical and materials systems using a multi-task approach, DPA-2 demonstrates superior generalization capabilities across multiple downstream tasks compared to the traditional single-task pre-training and fine-tuning methodologies. Our approach sets the stage for the development and broad application of LAMs in molecular and materials simulation research. Read More View Full Article |
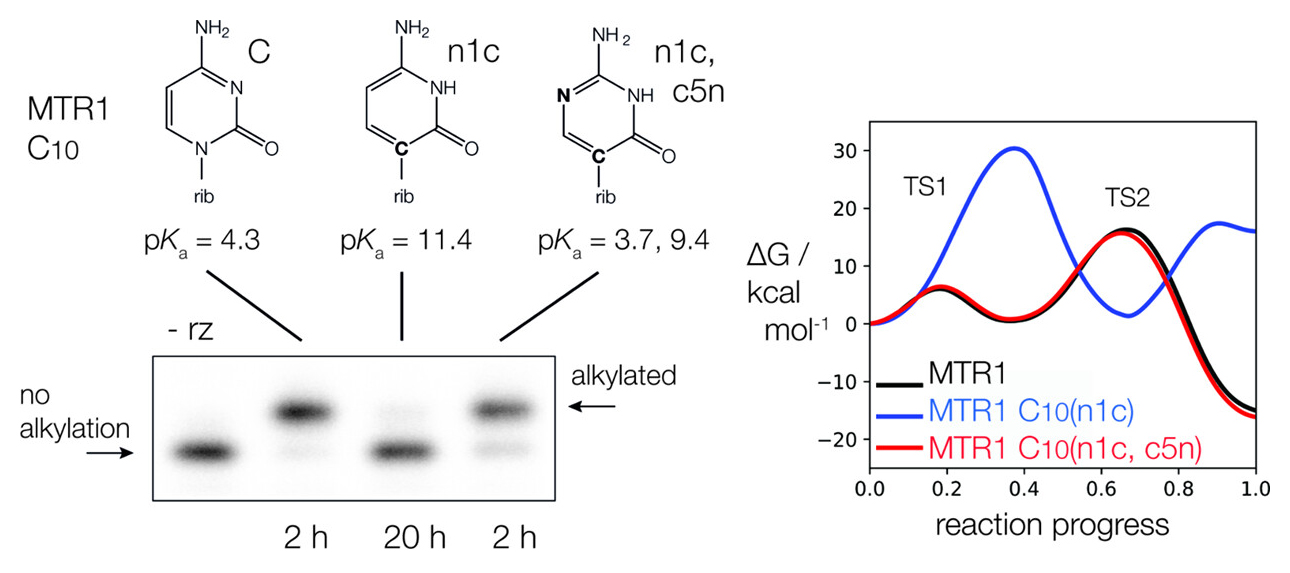 | The Role of General Acid Catalysis in the Mechanism of an Alkyl Transferase Ribozyme ACS Catalysis (2024) 14, 15294-15305 DOI: acscatal.4c04571 MTR1 is an in vitro-selected alkyl transferase ribozyme that transfers an alkyl group from O6-alkylguanine to N1 of the target adenine in the RNA substrate (A63). The structure of the ribozyme suggested a mechanism in which a cytosine (C10) acts as a general acid to protonate O6-alkylguanine N1. Here, we have analyzed the role of the C10 general acid and the A63 nucleophile by atomic mutagenesis and computation. C10 was substituted by n1c and n1c, c5n variants. The n1c variant has an elevated pKa (11.4 as the free nucleotide) and leads to a 104-fold lower activity that is pH-independent. Addition of the second c5n substitution with a lower pKa restored both the rate and pH dependence of alkyl transfer. Quantum mechanical calculations indicate that protonation of O6-alkylguanine lowers the barrier to alkyl transfer and that there is a significantly elevated barrier to proton transfer for the n1c single substitution. The calculated pKa values are in good agreement with the apparent values from measured rates. Increasing the pKa of the nucleophile by A63 n7c substitution led to a 6-fold higher rate. The increased reactivity of the nucleophile corresponds to a βnuc of ∼0.5, indicating significant C–N bond formation in the transition state. Taken together, these results are consistent with a two-step mechanism comprising protonation of the O6-alkylguanine followed by alkyl transfer. Read More View Full Article Download PDF |
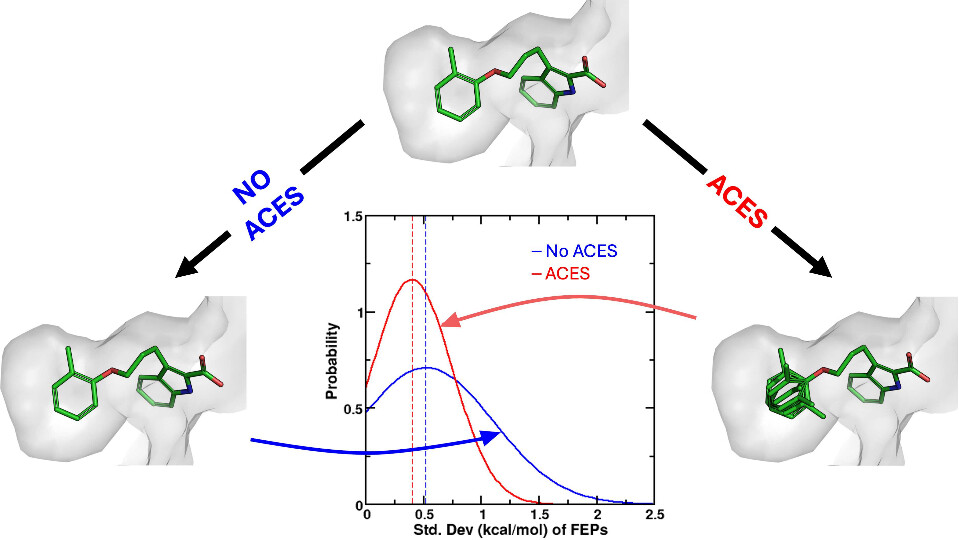 | Improvements in Precision of Relative Binding Free Energy Calculations Afforded by the Alchemical Enhanced Sampling (ACES) Approach Journal of Chemical Information and Modeling (2024) 64, 7046-7055 DOI: 10.1021/acs.jcim.4c00464 Accurate in silico predictions of how strongly small molecules bind to proteins, such as those afforded by relative binding free energy (RBFE) calculations, can greatly increase the efficiency of the hit-to-lead and lead optimization stages of the drug discovery process. The success of such calculations, however, relies heavily on their precision. Here, we show that a recently developed alchemical enhanced sampling (ACES) approach can consistently improve the precision of RBFE calculations on a large and diverse set of proteins and small molecule ligands. The addition of ACES to conventional RBFE calculations lowered the average hysteresis by over 35% (0.3–0.4 kcal/mol) and the average replicate spread by over 25% (0.2–0.3 kcal/mol) across a set of 10 protein targets and 213 small molecules while maintaining similar or improved accuracy. We show in atomic detail how ACES improved convergence of several representative RBFE calculations through enhancing the sampling of important slowly transitioning ligand degrees of freedom. Read More View Full Article Download PDF |
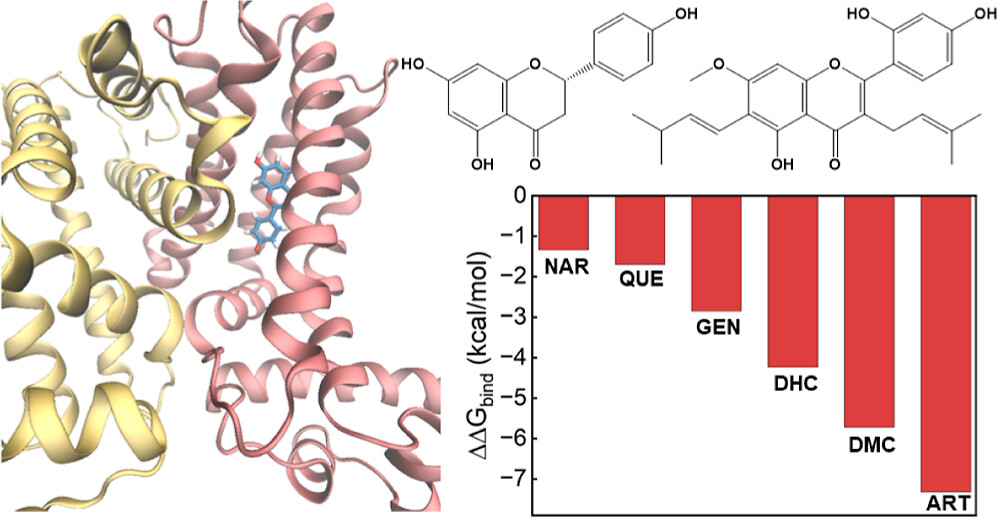 | Adsorption of Flavonoids in a Transcriptional Regulator TtgR: Relative Binding Free Energies and Intermolecular Interactions The Journal of Physical Chemistry B (2024) 128, 6529-6541 DOI: 10.1021/acs.jpcb.4c02303 Antimicrobial resistance in bacteria often arises from their ability to actively identify and expel toxic compounds. The bacterium strain Pseudomonas putida DOT-T1E utilizes its TtgABC efflux pump to confer robust resistance against antibiotics, flavonoids, and organic solvents. This resistance mechanism is intricately regulated at the transcriptional level by the TtgR protein. Through molecular dynamics and alchemical free energy simulations, we systematically examine the binding of seven flavonoids and their derivatives with the TtgR transcriptional regulator. Our simulations reveal distinct binding geometries and free energies for the flavonoids in the active site of the protein, which are driven by a range of noncovalent forces encompassing van der Waals, electrostatic, and hydrogen bonding interactions. The interplay of molecular structures, substituent patterns, and intermolecular interactions effectively stabilizes the bound flavonoids, confining their movements within the TtgR binding pocket. These findings yield valuable insights into the molecular determinants that govern ligand recognition in TtgR and shed light on the mechanism of antimicrobial resistance in P. putida DOT-T1E. Read More View Full Article Download PDF |
 | Software Infrastructure for Next-Generation QM/MM−ΔMLP Force Fields The Journal of Physical Chemistry B (2024) 128, 6257-6271 DOI: 10.1021/acs.jpcb.4c01466 We present software infrastructure for the design and testing of new quantum mechanical/molecular mechanical and machine-learning potential (QM/MM−ΔMLP) force fields for a wide range of applications. The software integrates Amber’s molecular dynamics simulation capabilities with fast, approximate quantum models in the xtb package and machine-learning potential corrections in DeePMD-kit. The xtb package implements the recently developed density-functional tight-binding QM models with multipolar electrostatics and density-dependent dispersion (GFN2-xTB), and the interface with Amber enables their use in periodic boundary QM/MM simulations with linear-scaling QM/MM particle-mesh Ewald electrostatics. The accuracy of the semiempirical models is enhanced by including machine-learning correction potentials (ΔMLPs) enabled through an interface with the DeePMD-kit software. The goal of this paper is to present and validate the implementation of this software infrastructure in molecular dynamics and free energy simulations. The utility of the new infrastructure is demonstrated in proof-of-concept example applications. The software elements presented here are open source and freely available. Their interface provides a powerful enabling technology for the design of new QM/MM−ΔMLP models for studying a wide range of problems, including biomolecular reactivity and protein–ligand binding. Read More View Full Article Download PDF |
 | Amber free energy tools: Interoperable software for free energy simulations using generalized quantum mechanical/molecular mechanical and machine learning potentials The Journal of Chemical Physics (2024) 160, 224104 DOI: 10.1063/5.0211276 We report the development and testing of new integrated cyberinfrastructure for performing free energy simulations with generalized hybrid quantum mechanical/molecular mechanical (QM/MM) and machine learning potentials (MLPs) in Amber. The Sander molecular dynamics program has been extended to leverage fast, density-functional tight-binding models implemented in the DFTB+ and xTB packages, and an interface to the DeePMD-kit software enables the use of MLPs. The software is integrated through application program interfaces that circumvent the need to perform “system calls” and enable the incorporation of long-range Ewald electrostatics into the external software’s self-consistent field procedure. The infrastructure provides access to QM/MM models that may serve as the foundation for QM/MM–ΔMLP potentials, which supplement the semiempirical QM/MM model with a MLP correction trained to reproduce ab initio QM/MM energies and forces. Efficient optimization of minimum free energy pathways is enabled through a new surface-accelerated finite-temperature string method implemented in the FE-ToolKit package. Furthermore, we interfaced Sander with the i-PI software by implementing the socket communication protocol used in the i-PI client–server model. The new interface with i-PI allows for the treatment of nuclear quantum effects with semiempirical QM/MM–ΔMLP models. The modular interoperable software is demonstrated on proton transfer reactions in guanine-thymine mispairs in a B-form deoxyribonucleic acid helix. The current work represents a considerable advance in the development of modular software for performing free energy simulations of chemical reactions that are important in a wide range of applications. Read More View Full Article Download PDF |
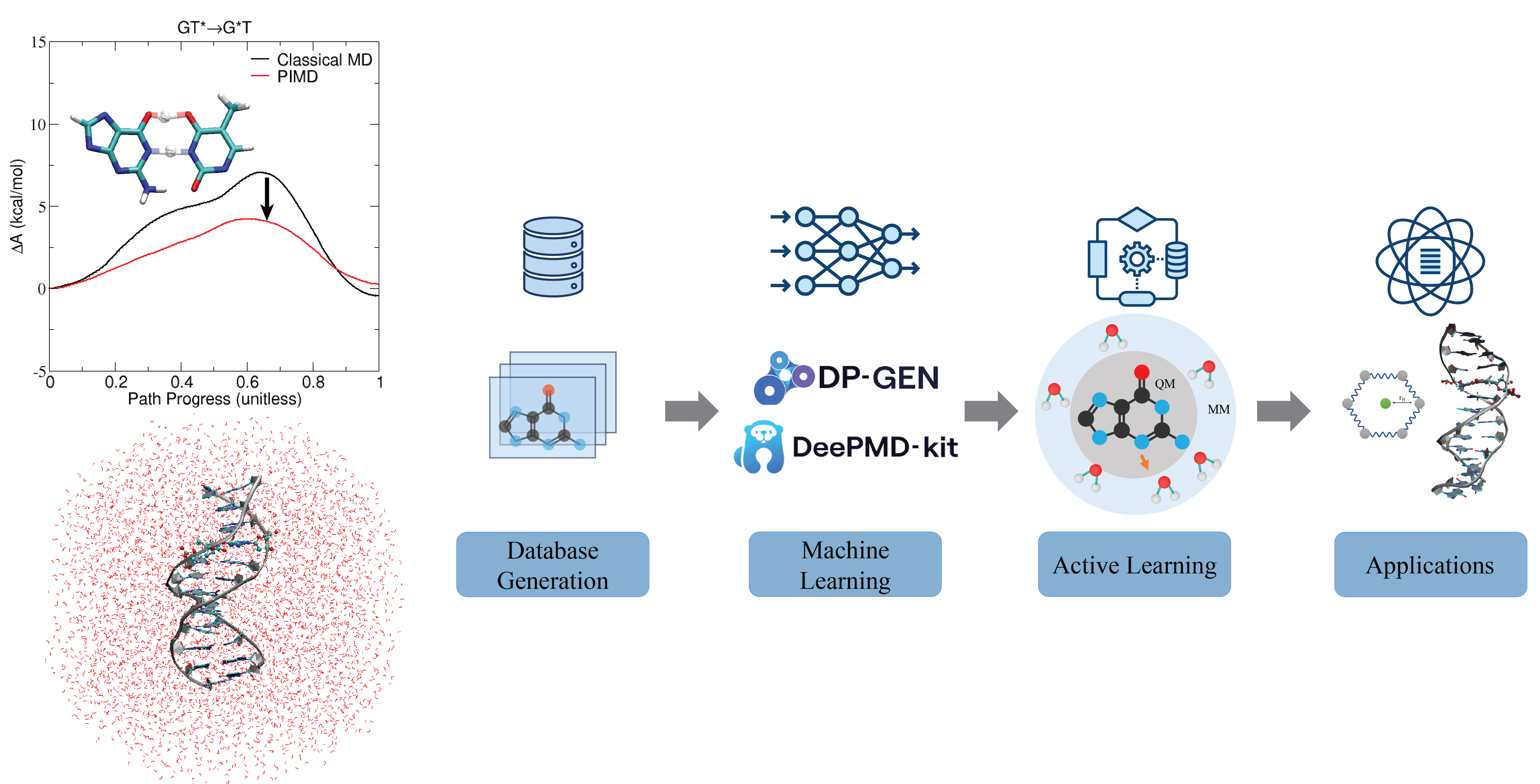 | Electronic and Nuclear Quantum Effects on Proton Transfer Reactions of Guanine–Thymine (G-T) Mispairs Using Combined Quantum Mechanical/Molecular Mechanical and Machine Learning Potentials Molecules (2024) 29, 2703 DOI: 10.3390/molecules29112703 Rare tautomeric forms of nucleobases can lead to Watson–Crick-like (WC-like) mispairs in DNA, but the process of proton transfer is fast and difficult to detect experimentally. NMR studies show evidence for the existence of short-time WC-like guanine–thymine (G-T) mispairs; however, the mechanism of proton transfer and the degree to which nuclear quantum effects play a role are unclear. We use a B-DNA helix exhibiting a wGT mispair as a model system to study tautomerization reactions. We perform ab initio (PBE0/6-31G*) quantum mechanical/molecular mechanical (QM/MM) simulations to examine the free energy surface for tautomerization. We demonstrate that while the ab initio QM/MM simulations are accurate, considerable sampling is required to achieve high precision in the free energy barriers. To address this problem, we develop a QM/MM machine learning potential correction (QM/MM-ΔMLP) that is able to improve the computational efficiency, greatly extend the accessible time scales of the simulations, and enable practical application of path integral molecular dynamics to examine nuclear quantum effects. We find that the inclusion of nuclear quantum effects has only a modest effect on the mechanistic pathway but leads to a considerable lowering of the free energy barrier for the GT*⇌G*T equilibrium. Our results enable a rationalization of observed experimental data and the prediction of populations of rare tautomeric forms of nucleobases and rates of their interconversion in B-DNA. Read More View Full Article Download PDF |
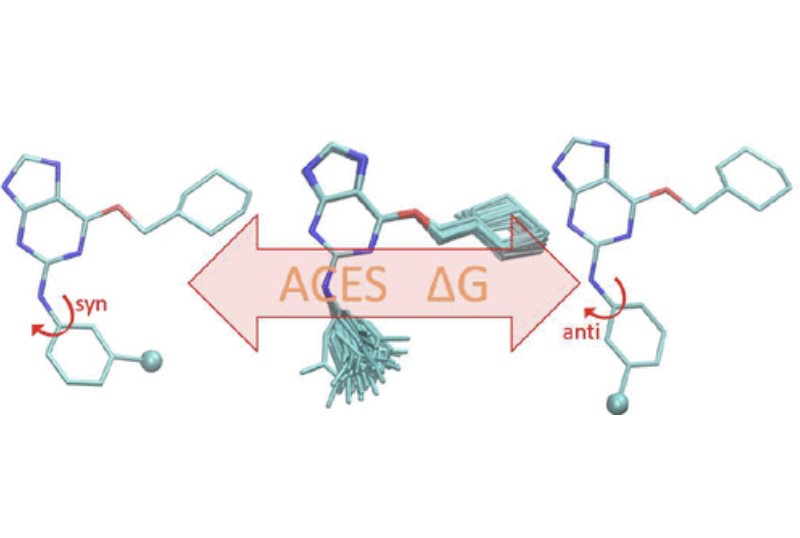
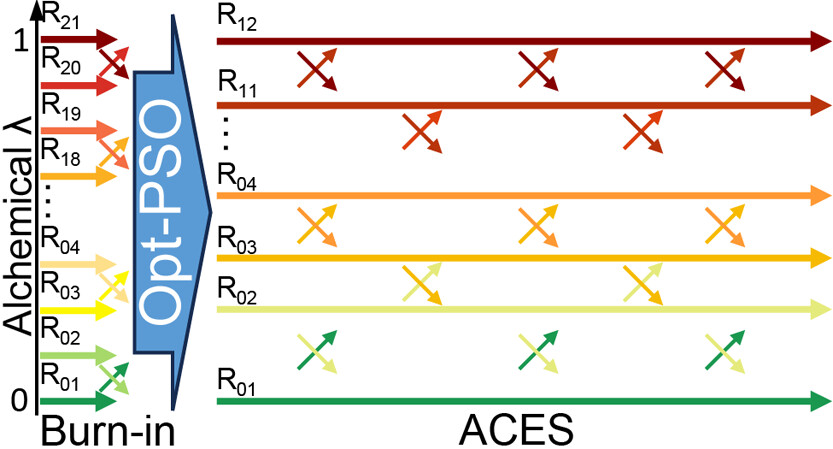 | Alchemical Enhanced Sampling with Optimized Phase Space Overlap Journal of Chemical Theory and Computation (2024) 20, 3935-3953 DOI: doi.org/10.1021/acs.jctc.4c00251 An alchemical enhanced sampling (ACES) method has recently been introduced to facilitate importance sampling in free energy simulations. The method achieves enhanced sampling from Hamiltonian replica exchange within a dual topology framework while utilizing new smoothstep softcore potentials. A common sampling problem encountered in lead optimization is the functionalization of aromatic rings that exhibit distinct conformational preferences when interacting with the protein. It is difficult to converge the distribution of ring conformations due to the long time scale of ring flipping events; however, the ACES method addresses this issue by modeling the syn and anti ring conformations within a dual topology. ACES thereby samples the conformer distributions by alchemically tunneling between states, as opposed to traversing a physical pathway with a high rotational barrier. We demonstrate the use of ACES to overcome conformational sampling issues involving ring flipping in ML300-derived noncovalent inhibitors of SARS-CoV-2 Main Protease (Mpro). The demonstrations explore how the use of replica exchange and the choice of softcore selection affects the convergence of the ring conformation distributions. Furthermore, we examine how the accuracy of the calculated free energies is affected by the degree of phase space overlap (PSO) between adjacent states (i.e., between neighboring λ-windows) and the Hamiltonian replica exchange acceptance ratios. Both of these factors are sensitive to the spacing between the intermediate states. We introduce a new method for choosing a schedule of λ values. The method analyzes short “burn-in” simulations to construct a 2D map of the nonlocal PSO. The schedule is obtained by optimizing an alchemical pathway on the 2D map that equalizes the PSO between the λ intervals. The optimized phase space overlap λ-spacing method (Opt-PSO) leads to more numerous end-to-end single passes and round trips due to the correlation between PSO and Hamiltonian replica exchange acceptance ratios. The improved exchange statistics enhance the efficiency of ACES method. The method has been implemented into the FE-ToolKit software package, which is freely available. Read More View Full Article Download PDF |
 | Surface-Accelerated String Method for Locating Minimum Free Energy Paths Journal of Chemical Theory and Computation (2024) 20, 2058–2073 DOI: 10.1021/acs.jctc.3c01401 We present a surface-accelerated string method (SASM) to efficiently optimize low-dimensional reaction pathways from the sampling performed with expensive quantum mechanical/molecular mechanical (QM/MM) Hamiltonians. The SASM accelerates the convergence of the path using the aggregate sampling obtained from the current and previous string iterations, whereas approaches like the string method in collective variables (SMCV) or the modified string method in collective variables (MSMCV) update the path only from the sampling obtained from the current iteration. Furthermore, the SASM decouples the number of images used to perform sampling from the number of synthetic images used to represent the path. The path is optimized on the current best estimate of the free energy surface obtained from all available sampling, and the proposed set of new simulations is not restricted to being located along the optimized path. Instead, the umbrella potential placement is chosen to extend the range of the free energy surface and improve the quality of the free energy estimates near the path. In this manner, the SASM is shown to improve the exploration for a minimum free energy pathway in regions where the free energy surface is relatively flat. Furthermore, it improves the quality of the free energy profile when the string is discretized with too few images. We compare the SASM, SMCV, and MSMCV using 3 QM/MM applications: a ribozyme methyltransferase reaction using 2 reaction coordinates, the 2′-O-transphosphorylation reaction of Hammerhead ribozyme using 3 reaction coordinates, and a tautomeric reaction in B-DNA using 5 reaction coordinates. We show that SASM converges the paths using roughly 3 times less sampling than the SMCV and MSMCV methods. All three algorithms have been implemented in the FE-ToolKit package made freely available. Read More View Full Article Download PDF |
 | AmberTools Journal of Chemical Information and Modeling (2023) 63, 6183-6191 DOI: 10.1021/acs.jcim.3c01153 AmberTools is a free and open-source collection of programs used to set up, run, and analyze molecular simulations. The newer features contained within AmberTools23 are briefly described in this Application note. Read More View Full Article Download PDF |
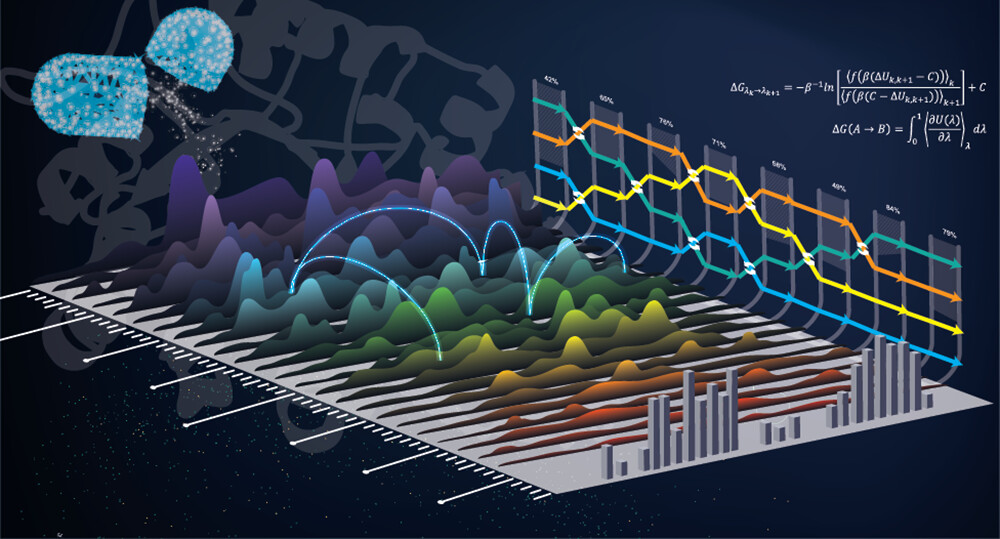 | Modern Alchemical Free Energy Methods for Drug Discovery Explained ACS Physical Chemistry Au (2023) 3, 478-491 DOI: 10.1021/acsphyschemau.3c00033 This Perspective provides a contextual explanation of the current state-of-the-art alchemical free energy methods and their role in drug discovery as well as highlights select emerging technologies. The narrative attempts to answer basic questions about what goes on “under the hood” in free energy simulations and provide general guidelines for how to run simulations and analyze the results. It is the hope that this work will provide a valuable introduction to students and scientists in the field. Read More View Full Article Download PDF |
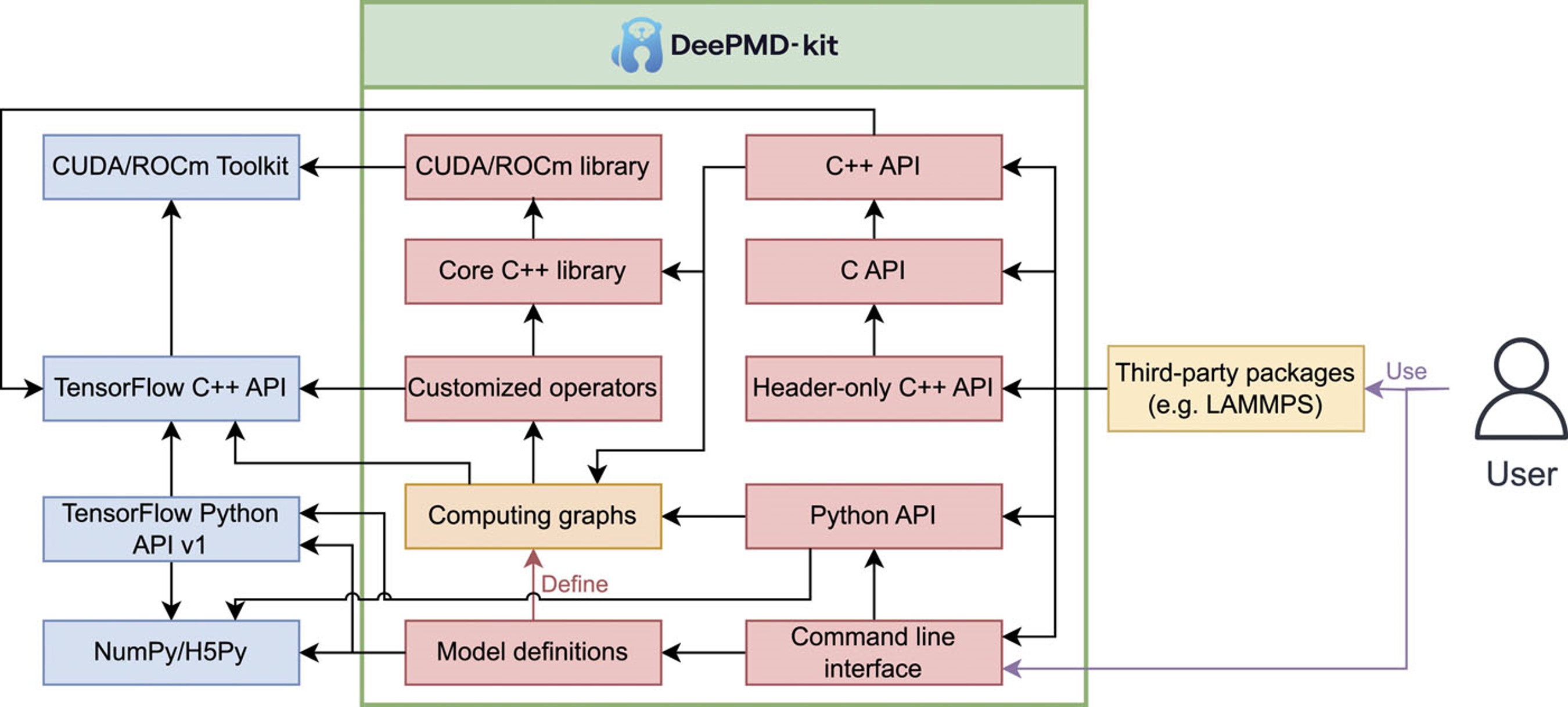 | DeePMD-kit v2: A software package for deep potential models The Journal of Chemical Physics (2023) 159, 054801 DOI: 10.1063/5.0155600 DeePMD-kit is a powerful open-source software package that facilitates molecular dynamics simulations using machine learning potentials known as Deep Potential (DP) models. This package, which was released in 2017, has been widely used in the fields of physics, chemistry, biology, and material science for studying atomistic systems. The current version of DeePMD-kit offers numerous advanced features, such as DeepPot-SE, attention-based and hybrid descriptors, the ability to fit tensile properties, type embedding, model deviation, DP-range correction, DP long range, graphics processing unit support for customized operators, model compression, non-von Neumann molecular dynamics, and improved usability, including documentation, compiled binary packages, graphical user interfaces, and application programming interfaces. This article presents an overview of the current major version of the DeePMD-kit package, highlighting its features and technical details. Additionally, this article presents a comprehensive procedure for conducting molecular dynamics as a representative application, benchmarks the accuracy and efficiency of different models, and discusses ongoing developments. Read More View Full Article Download PDF |
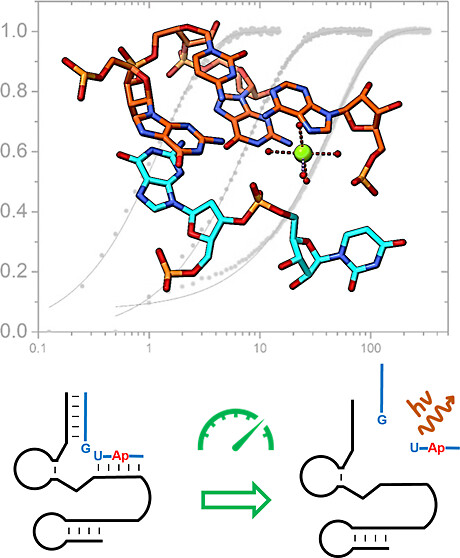 | Rapid Kinetics of Pistol Ribozyme: Insights into Limits to RNA Catalysis Biochemistry (2023) 62, 2079-2092 DOI: 10.1021/acs.biochem.3c00160 Pistol ribozyme (Psr) is a distinct class of small endonucleolytic ribozymes, which are important experimental systems for defining fundamental principles of RNA catalysis and designing valuable tools in biotechnology. High-resolution structures of Psr, extensive structure–function studies, and computation support a mechanism involving one or more catalytic guanosine nucleobases acting as a general base and divalent metal ion-bound water acting as an acid to catalyze RNA 2′-O-transphosphorylation. Yet, for a wide range of pH and metal ion concentrations, the rate of Psr catalysis is too fast to measure manually and the reaction steps that limit catalysis are not well understood. Here, we use stopped-flow fluorescence spectroscopy to evaluate Psr temperature dependence, solvent H/D isotope effects, and divalent metal ion affinity and specificity unconstrained by limitations due to fast kinetics. The results show that Psr catalysis is characterized by small apparent activation enthalpy and entropy changes and minimal transition state H/D fractionation, suggesting that one or more pre-equilibrium steps rather than chemistry is rate limiting. Quantitative analyses of divalent ion dependence confirm that metal aquo ion pKa correlates with higher rates of catalysis independent of differences in ion binding affinity. However, ambiguity regarding the rate-limiting step and similar correlation with related attributes such as ionic radius and hydration free energy complicate a definitive mechanistic interpretation. These new data provide a framework for further interrogation of Psr transition state stabilization and show how thermal instability, metal ion insolubility at optimal pH, and pre-equilibrium steps such as ion binding and folding limit the catalytic power of Psr suggesting potential strategies for further optimization. Read More View Full Article Download PDF |
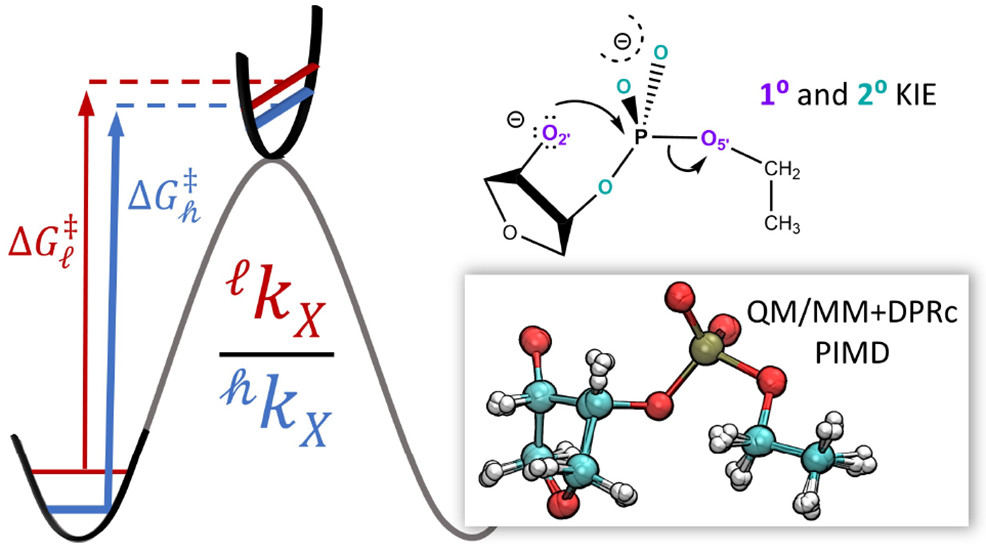
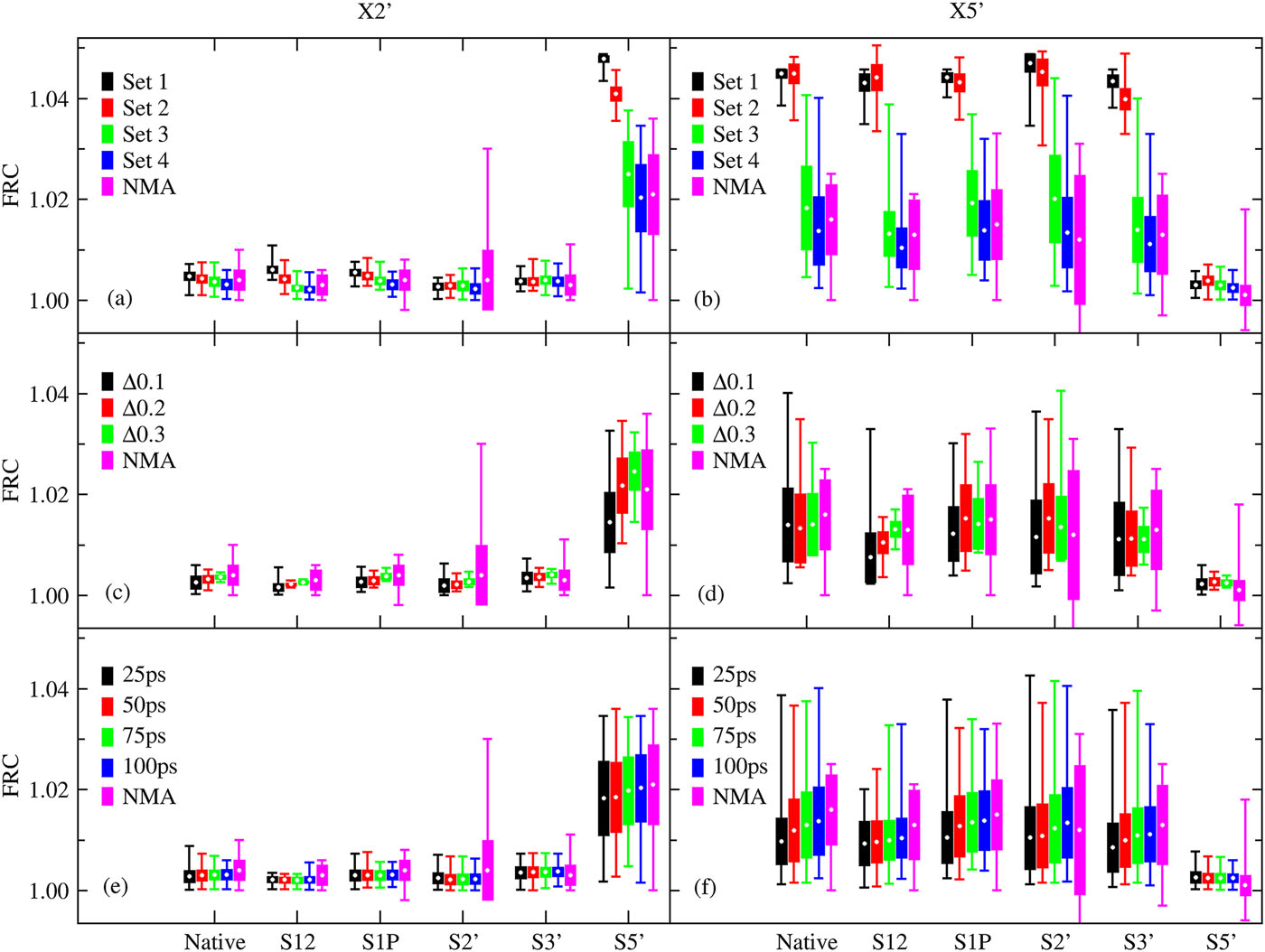 | Estimation of frequency factors for the calculation of kinetic isotope effects from classical and path integral free energy simulations The Journal of Chemical Physics (2023) 158, 174105-174115 DOI: 10.1063/5.0147218 We use the modified Bigeleisen–Mayer equation to compute kinetic isotope effect values for non-enzymatic phosphoryl transfer reactions from classical and path integral molecular dynamics umbrella sampling. The modified form of the Bigeleisen–Mayer equation consists of a ratio of imaginary mode vibrational frequencies and a contribution arising from the isotopic substitution’s effect on the activation free energy, which can be computed from path integral simulation. In the present study, we describe a practical method for estimating the frequency ratio correction directly from umbrella sampling in a manner that does not require normal mode analysis of many geometry optimized structures. Instead, the method relates the frequency ratio to the change in the mass weighted coordinate representation of the minimum free energy path at the transition state induced by isotopic substitution. The method is applied to the calculation of 16/18O and 32/34S primary kinetic isotope effect values for six non-enzymatic phosphoryl transfer reactions. We demonstrate that the results are consistent with the analysis of geometry optimized transition state ensembles using the traditional Bigeleisen–Mayer equation. The method thus presents a new practical tool to enable facile calculation of kinetic isotope effect values for complex chemical reactions in the condensed phase. Read More View Full Article Download PDF |
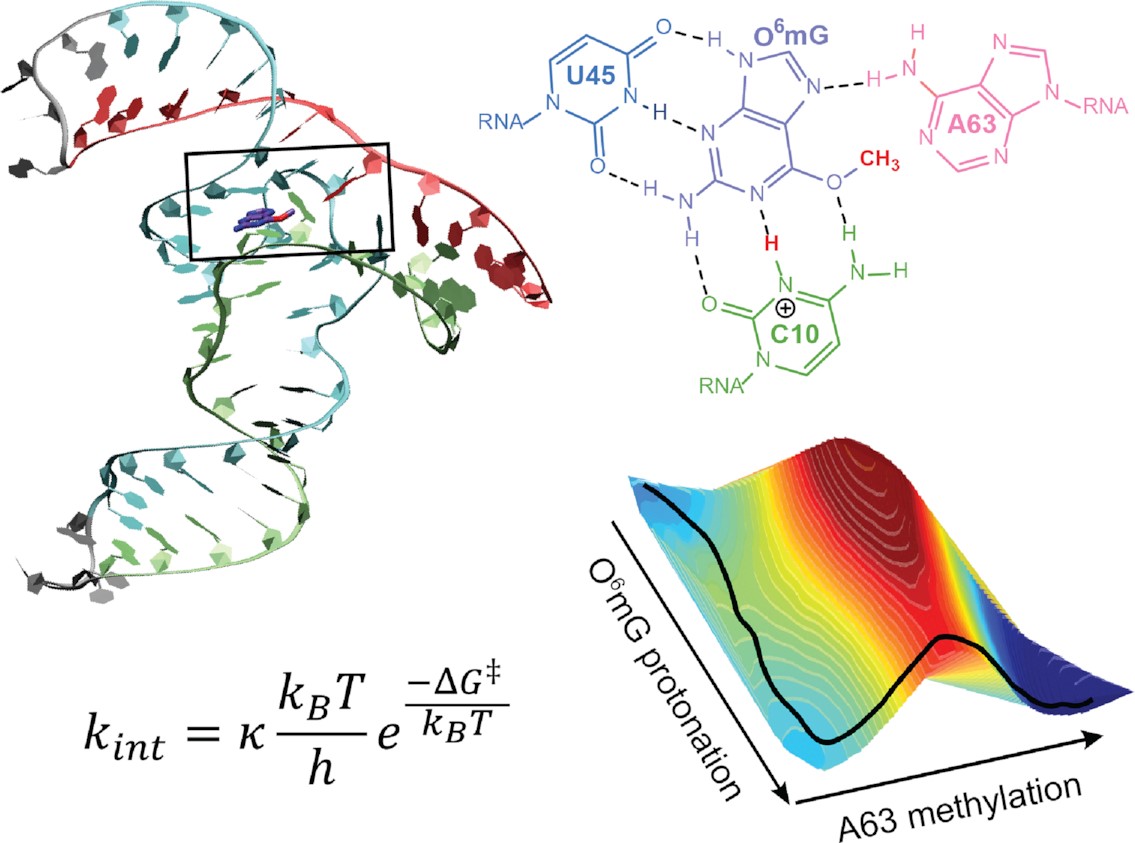 | Catalytic mechanism and pH dependence of a methyltransferase ribozyme (MTR1) from computational enzymology Nucleic Acids Research (2023) 51, 4508-4518 DOI: 10.1093/nar/gkad260 A methyltransferase ribozyme (MTR1) was selected in vitro to catalyze alkyl transfer from exogenous O6-methylguanine (O6mG) to a target adenine N1, and recently, high-resolution crystal structures have become available. We use a combination of classical molecular dynamics, ab initio quantum mechanical/molecular mechanical (QM/MM) and alchemical free energy (AFE) simulations to elucidate the atomic-level solution mechanism of MTR1. Simulations identify an active reactant state involving protonation of C10 that hydrogen bonds with O6mG:N1. The deduced mechanism involves a stepwise mechanism with two transition states corresponding to proton transfer from C10:N3 to O6mG:N1 and rate-controlling methyl transfer (19.4 kcal·mol−1 barrier). AFE simulations predict the pKa for C10 to be 6.3, close to the experimental apparent pKa of 6.2, further implicating it as a critical general acid. The intrinsic rate derived from QM/MM simulations, together with pKa calculations, enables us to predict an activity–pH profile that agrees well with experiment. The insights gained provide further support for a putative RNA world and establish new design principles for RNA-based biochemical tools. Read More View Full Article Download PDF |
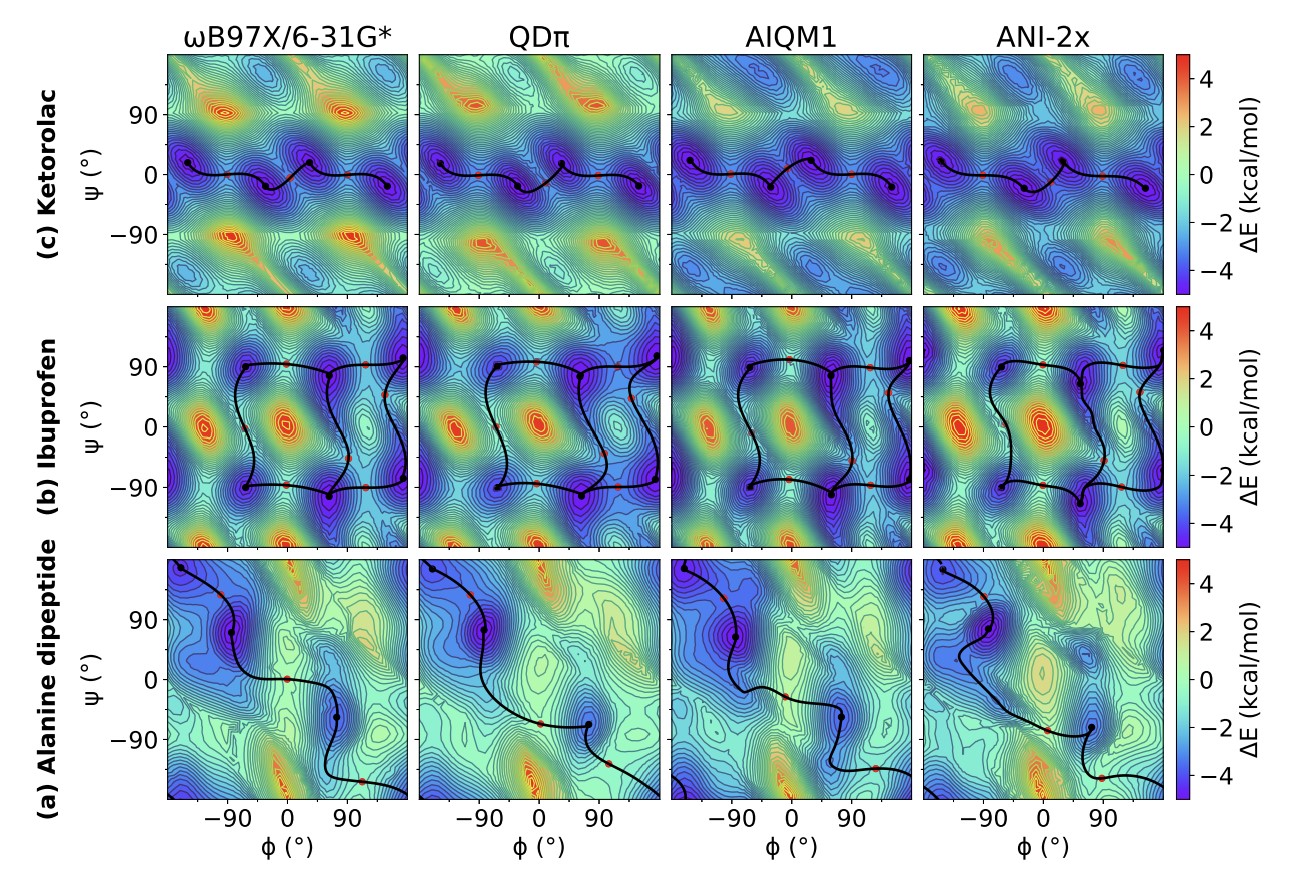 | Modern semiempirical electronic structure methods and machine learning potentials for drug discovery: conformers, tautomers and protonation states The Journal of Chemical Physics (2023) 158, 124110 DOI: 10.1063/5.0139281 Modern semiempirical electronic structure methods have considerable promise in drug discovery as universal "force fields" that can reliably model biological and drug-like molecules. Herein, we compare the performance of several NDDO-based semiempirical (MNDO/d, AM1, PM6 and ODM2), density-functional tight-binding based (DFTB3, GFN1-xTB and GFN2-xTB) models with pure machine learning potentials (ANI-1x and ANI-2x) and hybrid quantum mechanical/machine learning potentials (AIQM1 and QDπ) for a wide range of data computed at a consistent ωB97X/6-31G* level of theory (as in the ANI-1x database). This data includes conformational energies, intermolecular interactions, tautomers, and protonation states. Additional comparisons are made to a set of natural and synthetic nucleic acids from the artificially expanded genetic information system (AEGIS). This dataset has important implications in the design of new biotechnology and therapeutics. Finally, weexamine acid/base chemistry relevant for RNA cleavage reactions catalyzed by small nucleolytic ribozymes and ribonucleases. Overall, the recently developed QDπ model performs exceptionally well across all datasets, having especially high accuracy for tautomers and protonation states relevant to drug discovery. Read More View Full Article Download PDF |
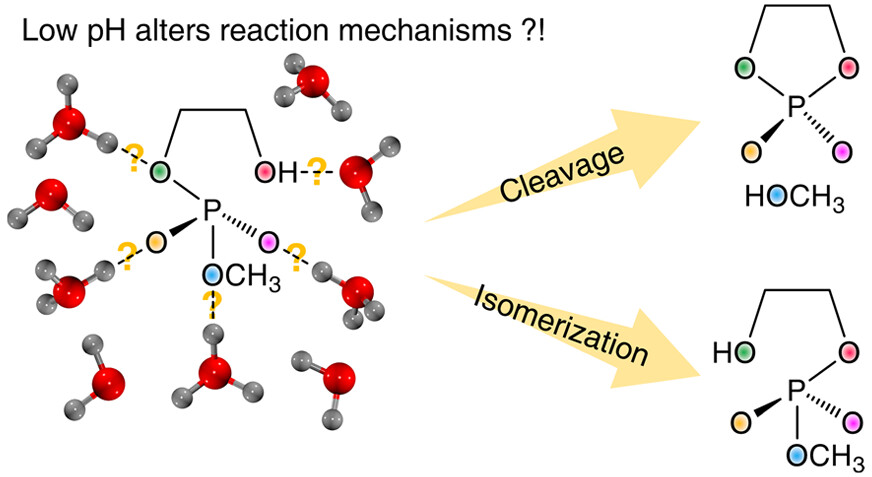 | Altered Mechanisms for Acid-Catalyzed RNA Cleavage and Isomerization Reactions Models Journal of Chemical Theory and Computation (2023) 19, 1322-1332 DOI: 10.1021/acs.jctc.2c01277 RNA strand cleavage by 2′-O-transphosphorylation is catalyzed not only by numerous nucleolytic RNA enzymes (ribozymes) but also by hydroxide or hydronium ions. In experiments, both cleavage of the 5′-linked nucleoside and isomerization between 3′,5′- and 2′,5′-phosphodiesters occur under acidic conditions, while only the cleavage reaction is observed under basic conditions. An ab initio path-integral approach for simulating kinetic isotope effects is used to reveal the reaction mechanisms for RNA cleavage and isomerization reactions under acidic conditions. Moreover, the proposed mechanisms can also be combined through the experimental pH-rate profiles. Read More View Full Article Download PDF |
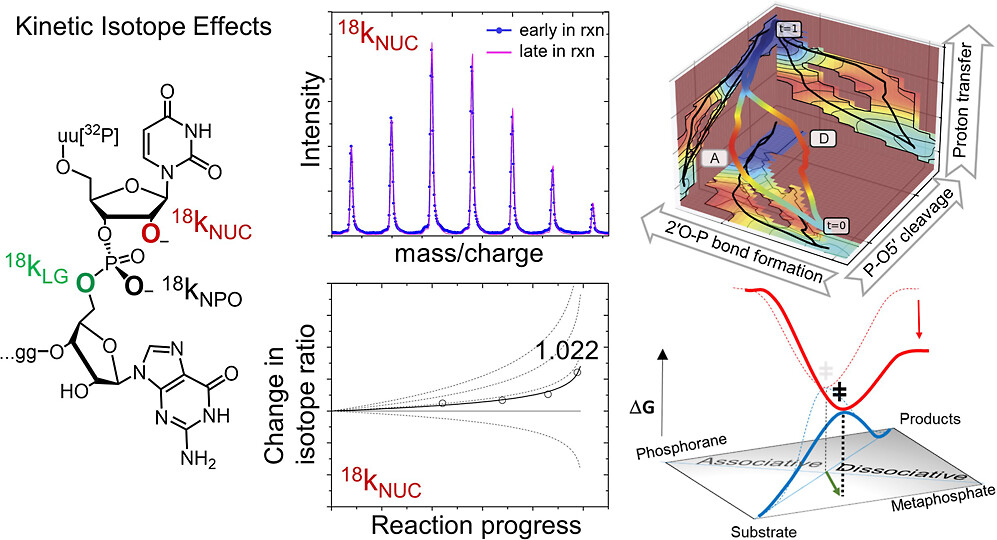 | Dissociative Transition State in Hepatitis Delta Virus Ribozyme Catalysis Journal of the American Chemical Society (2023) 145, 2830-2839 DOI: 10.1021/jacs.2c10079 Ribonucleases and small nucleolytic ribozymes are both able to catalyze RNA strand cleavage through 2′-O-transphosphorylation, provoking the question of whether protein and RNA enzymes facilitate mechanisms that pass through the same or distinct transition states. Here, we report the primary and secondary 18O kinetic isotope effects for hepatitis delta virus ribozyme catalysis that reveal a dissociative, metaphosphate-like transition state in stark contrast to the late, associative transition states observed for reactions catalyzed by specific base, Zn2+ ions, or ribonuclease A. This new information provides evidence for a discrete ribozyme active site design that modulates the RNA cleavage pathway to pass through an altered transition state. Read More View Full Article Download PDF |
 | Chapter 6 Learning DeePMD-Kit: A Guide to Building Deep Potential Models In A Practical Guide to Recent Advances in Multiscale Modeling and Simulation of Biomolecules (2023) Chapter: 6 Edited by: Yong Wang and Ruhong Zhou Publishers: AIP Publishing DOI: 10.1063/9780735425279_006 ISBN: 978-0-7354-2527-9 A new direction has emerged in molecular simulations in recent years, where potential energy surfaces (PES) are constructed using machine learning (ML) methods. These ML models, combining the accuracy of quantum mechanical models and the efficiency of empirical atomic potential models, have been demonstrated by many studies to have extensive application prospects. This chapter introduces a recently developed ML model, Deep Potential (DP), and the corresponding package, DeePMD-kit. First, we present the basic theory of the DP method. Then, we show how to train and test a DP model for a gas-phase methane molecule using the DeePMD-kit package. Next, we introduce some recent progress on simulations of biomolecular processes by integrating the DeePMD-kit with the AMBER molecular simulation software suite. Finally, we provide a supplement on points that require further explanation. Read More View Full Article Download PDF |
 | QDπ: A Quantum Deep Potential Interaction Model for Drug Discovery Journal of Chemical Theory and Computation (2023) 19, 1261-1275 DOI: 10.1021/acs.jctc.2c01172 We report QDπ-v1.0 for modeling the internal energy of drug molecules containing H, C, N, and O atoms. The QDπ model is in the form of a quantum mechanical/machine learning potential correction (QM/Δ-MLP) that uses a fast third-order self-consistent density-functional tight-binding (DFTB3/3OB) model that is corrected to a quantitatively high-level of accuracy through a deep-learning potential (DeepPot-SE). The model has the advantage that it is able to properly treat electrostatic interactions and handle changes in charge/protonation states. The model is trained against reference data computed at the ωB97X/6-31G* level (as in the ANI-1x data set) and compared to several other approximate semiempirical and machine learning potentials (ANI-1x, ANI-2x, DFTB3, MNDO/d, AM1, PM6, GFN1-xTB, and GFN2-xTB). The QDπ model is demonstrated to be accurate for a wide range of intra- and intermolecular interactions (despite its intended use as an internal energy model) and has shown to perform exceptionally well for relative protonation/deprotonation energies and tautomers. An example application to model reactions involved in RNA strand cleavage catalyzed by protein and nucleic acid enzymes illustrates QDπ has average errors less than 0.5 kcal/mol, whereas the other models compared have errors over an order of magnitude greater. Taken together, this makes QDπ highly attractive as a potential force field model for drug discovery. Read More View Full Article Download PDF |
 | ACES: Optimized Alchemically Enhanced Sampling Journal of Chemical Theory and Computation (2023) 19, 472-487 DOI: 10.1021/acs.jctc.2c00697 We present an alchemical enhanced sampling (ACES) method implemented in the GPU-accelerated AMBER free energy MD engine. The methods hinges on the creation of an “enhanced sampling state” by reducing or eliminating selected potential energy terms and interactions that lead to kinetic traps and conformational barriers while maintaining those terms that curtail the need to otherwise sample large volumes of phase space. For example, the enhanced sampling state might involve transforming regions of a ligand and/or protein side chain into a noninteracting “dummy state” with internal electrostatics and torsion angle terms turned off. The enhanced sampling state is connected to a real-state end point through a Hamiltonian replica exchange (HREMD) framework that is facilitated by newly developed alchemical transformation pathways and smoothstep softcore potentials. This creates a counterdiffusion of real and enhanced-sampling states along the HREMD network. The effect of a differential response of the environment to the real and enhanced-sampling states is minimized by leveraging the dual topology framework in AMBER to construct a counterbalancing HREMD network in the opposite alchemical direction with the same (or similar) real and enhanced sampling states at inverted end points. The method has been demonstrated in a series of test cases of increasing complexity where traditional MD, and in several cases alternative REST2-like enhanced sampling methods, are shown to fail. The hydration free energy for acetic acid was shown to be independent of the starting conformation, and the values for four additional edge case molecules from the FreeSolv database were shown to have a significantly closer agreement with experiment using ACES. The method was further able to handle different rotamer states in a Cdk2 ligand identified as fractionally occupied in crystal structures. Finally, ACES was applied to T4-lysozyme and demonstrated that the side chain distribution of V111χ1 could be reliably reproduced for the apo state, bound to p-xylene, and in p-xylene→ benzene transformations. In these cases, the ACES method is shown to be highly robust and superior to a REST2-like enhanced sampling implementation alone. Read More View Full Article Download PDF |
 | AMBER Free Energy Tools: A New Framework for the Design of Optimized Alchemical Transformation Pathways Journal of Chemical Theory and Computation (2023) 19, 640-658 DOI: 10.1021/acs.jctc.2c00725 We develop a framework for the design of optimized alchemical transformation pathways in free energy simulations using nonlinear mixing and a new functional form for so-called “softcore” potentials. We describe the implementation and testing of this framework in the GPU-accelerated AMBER software suite. The new optimized alchemical transformation pathways integrate a number of important features, including (1) the use of smoothstep functions to stabilize behavior near the transformation end points, (2) consistent power scaling of Coulomb and Lennard-Jones (LJ) interactions with unitless control parameters to maintain balance of electrostatic attractions and exchange repulsions, (3) pairwise form based on the LJ contact radius for the effective interaction distance with separation-shifted scaling, and (4) rigorous smoothing of the potential at the nonbonded cutoff boundary. The new softcore potential form is combined with smoothly transforming nonlinear λ weights for mixing specific potential energy terms, along with flexible λ-scheduling features, to enable robust and stable alchemical transformation pathways. The resulting pathways are demonstrated and tested, and shown to be superior to the traditional methods in terms of numerical stability and minimal variance of the free energy estimates for all cases considered. The framework presented here can be used to design new alchemical enhanced sampling methods, and leveraged in robust free energy workflows for large ligand data sets. Read More View Full Article Download PDF |
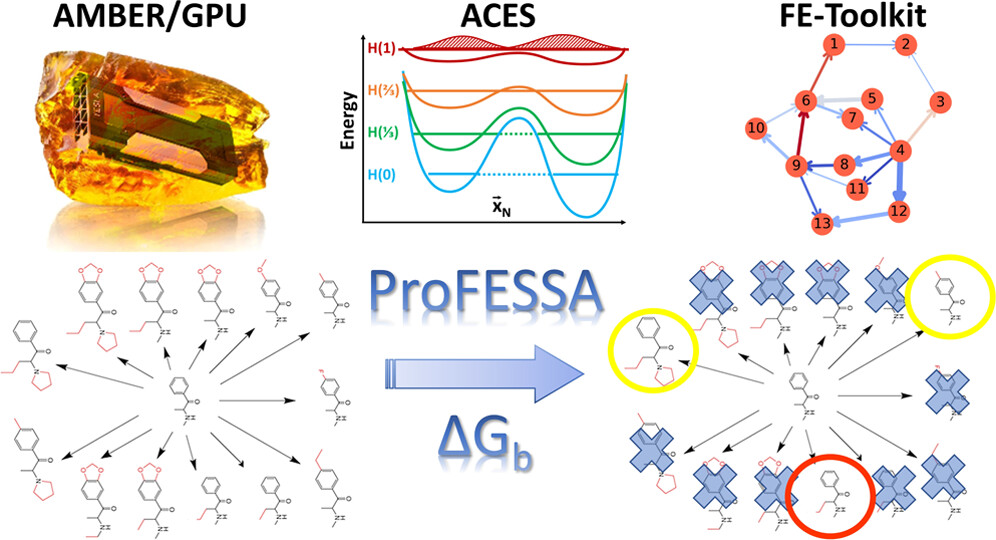 | AMBER Drug Discovery Boost Tools: Automated Workflow for Production Free-Energy Simulation Setup and Analysis (ProFESSA) Journal of Chemical Information and Modeling (2022) 62, 6069-6083 DOI: 10.1021/acs.jcim.2c00879 We report an automated workflow for production free-energy simulation setup and analysis (ProFESSA) using the GPU-accelerated AMBER free-energy engine with enhanced sampling features and analysis tools, part of the AMBER Drug Discovery Boost package that has been integrated into the AMBER22 release. The workflow establishes a flexible, end-to-end pipeline for performing alchemical free-energy simulations that brings to bear technologies, including new enhanced sampling features and analysis tools, to practical drug discovery problems. ProFESSA provides the user with top-level control of large sets of free-energy calculations and offers access to the following key functionalities: (1) automated setup of file infrastructure; (2) enhanced conformational and alchemical sampling with the ACES method; and (3) network-wide free-energy analysis with the optional imposition of cycle closure and experimental constraints. The workflow is applied to perform absolute and relative solvation free-energy and relative ligand–protein binding free-energy calculations using different atom-mapping procedures. Results demonstrate that the workflow is internally consistent and highly robust. Further, the application of a new network-wide Lagrange multiplier constraint analysis that imposes key experimental constraints substantially improves binding free-energy predictions. Read More View Full Article Download PDF |
 | Multireference Generalization of the Weighted Thermodynamic Perturbation Method The Journal of Physical Chemistry A (2022) 126, 8519-8533 DOI: 10.1021/acs.jpca.2c06201 We describe the generalized weighted thermodynamic perturbation (gwTP) method for estimating the free energy surface of an expensive “high-level” potential energy function from the umbrella sampling performed with multiple inexpensive “low-level” reference potentials. The gwTP method is a generalization of the weighted thermodynamic perturbation (wTP) method developed by Li and co-workers [J. Chem. Theory Comput. 2018, 14, 5583–5596] that uses a single “low-level” reference potential. The gwTP method offers new possibilities in model design whereby the sampling generated from several low-level potentials may be combined (e.g., specific reaction parameter models that might have variable accuracy at different stages of a multistep reaction). The gwTP method is especially well suited for use with machine learning potentials (MLPs) that are trained against computationally expensive ab initio quantum mechanical/molecular mechanical (QM/MM) energies and forces using active learning procedures that naturally produce multiple distinct neural network potentials. Simulations can be performed with greater sampling using the fast MLPs and then corrected to the ab initio level using gwTP. The capabilities of the gwTP method are demonstrated by creating reference potentials based on the MNDO/d and DFTB2/MIO semiempirical models supplemented with the “range-corrected deep potential” (DPRc). The DPRc parameters are trained to ab initio QM/MM data, and the potentials are used to calculate the free energy surface of stepwise mechanisms for nonenzymatic RNA 2′-O-transesterification model reactions. The extended sampling made possible by the reference potentials allows one to identify unequilibrated portions of the simulations that are not always evident from the short time scale commonly used with ab initio QM/MM potentials. We show that the reference potential approach can yield more accurate ab initio free energy predictions than the wTP method or what can be reasonably afforded from explicit ab initio QM/MM sampling. Read More View Full Article Download PDF |
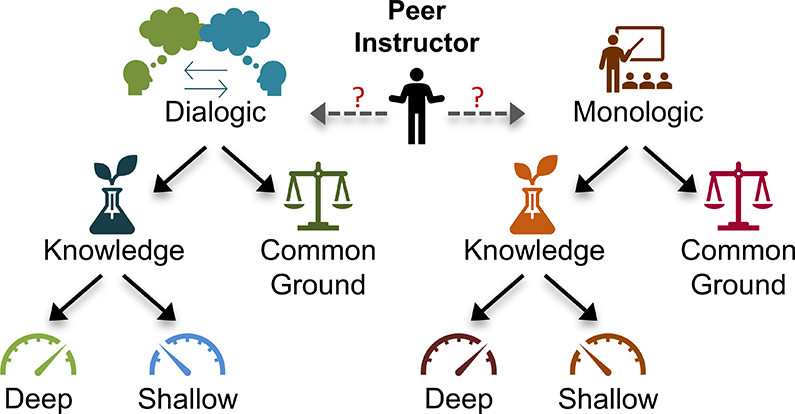 | Give and Take: Narrowing the Gap between Theory and Practice of Peer Instructors over Time Journal of Chemical Education (2022) 99, 3370-3385 DOI: 10.1021/acs.jchemed.2c00170 With the rise in implementation of peer-led learning in higher education, the interactions between peer instructors and their students warrant further investigation as an increasingly critical component of student learning. In this work, Teaching Interns (TIs) are undergraduate peer instructors that lead supplemental learning sessions in General Chemistry. Each week, TIs participate in pedagogy training and complete written reflections on their learning sessions. For this multicase study, six TIs were observed in their office hours over varying time periods. A qualitative approach was taken to analyze their verbal behaviors and the extent to which those behaviors matched their beliefs about teaching. Specifically, discourse analysis allowed for the characterization of the interactions between TIs and students, while analysis of TIs’ weekly written reflections provided insight into their teaching beliefs and perception of their own teaching sessions. The results presented here suggest that even at the start of the program, TIs hold some productive beliefs about teaching, though these beliefs were not always evident in their interactions with students. Over time, TIs generally shifted toward more student-centered discourse and honed their abilities to convey or elicit deeper knowledge among their students. Further, evidence from the TIs’ reflections suggest that they became better at self-monitoring their own teaching behaviors, shrinking the gap between their practices and espoused beliefs about teaching, and that they turned their focus toward student learning versus simply managing their sessions. Taken together, this work provides additional support for the further development and study of peer instruction programs. Read More View Full Article Download PDF |
 | RNA Electrostatics: How Ribozymes Engineer Active Sites to Enable Catalysis The Journal of Physical Chemistry B (2022) 126, 5982-5990 DOI: 10.1021/acs.jpcb.2c03727 Electrostatic interactions are fundamental to RNA structure and function, and intimately influenced by solvation and the ion atmosphere. RNA enzymes, or ribozymes, are catalytic RNAs that are able to enhance reaction rates over a million-fold, despite having only a limited repertoire of building blocks and available set of chemical functional groups. Ribozyme active sites usually occur at junctions where negatively charged helices come together, and in many cases leverage this strained electrostatic environment to recruit metal ions in solution that can assist in catalysis. Similar strategies have been implicated in related artificially engineered DNA enzymes. Herein, we apply Poisson–Boltzmann, 3D-RISM, and molecular simulations to study a set of metal-dependent small self-cleaving ribozymes (hammerhead, pistol, and Varkud satellite) as well as an artificially engineered DNAzyme (8–17) to examine electrostatic features and their relation to the recruitment of monovalent and divalent metal ions important for activity. We examine several fundamental roles for these ions that include: (1) structural integrity of the catalytically active state, (2) pKa tuning of residues involved in acid–base catalysis, and (3) direct electrostatic stabilization of the transition state via Lewis acid catalysis. Taken together, these examples demonstrate how RNA electrostatics orchestrates the site-specific and territorial binding of metal ions to play important roles in catalysis. Read More View Full Article Download PDF |
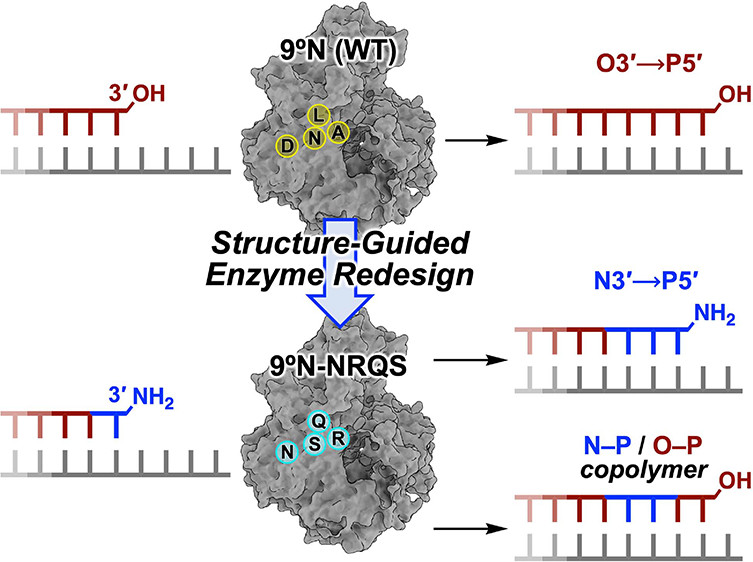 | Introducing a New Bond-Forming Activity in an Archaeal DNA Polymerase by Structure-Guided Enzyme Redesign ACS Chemical Biology (2022) 17, 1924-1936 DOI: 10.1021/acschembio.2c00373 PMC9442636 DNA polymerases have evolved to feature a highly conserved activity across the tree of life: formation of, without exception, internucleotidyl O–P linkages. Can this linkage selectivity be overcome by design to produce xenonucleic acids? Here, we report that the structure-guided redesign of an archaeal DNA polymerase, 9°N, exhibits a new activity undetectable in the wild-type enzyme: catalyzing the formation of internucleotidyl N–P linkages using 3′-NH2-ddNTPs. Replacing a metal-binding aspartate in the 9°N active site with asparagine was key to the emergence of this unnatural enzyme activity. MD simulations provided insights into how a single substitution enhances the productive positioning of a 3′-amino nucleophile in the active site. Further remodeling of the protein–nucleic acid interface in the finger subdomain yielded a quadruple-mutant variant (9°N-NRQS) displaying DNA-dependent NP-DNA polymerase activity. In addition, the engineered promiscuity of 9°N-NRQS was leveraged for one-pot synthesis of DNA─NP-DNA copolymers. This work sheds light on the molecular basis of substrate fidelity and latent promiscuity in enzymes. Read More View Full Article Download PDF |
 | Combined QM/MM, Machine Learning Path Integral Approach to Compute Free Energy Profiles and Kinetic Isotope Effects in RNA Cleavage Reactions Journal of Chemical Theory and Computation (2022) 18, 4304-4317 DOI: 10.1021/acs.jctc.2c00151 We present a fast, accurate, and robust approach for determination of free energy profiles and kinetic isotope effects for RNA 2′-O-transphosphorylation reactions with inclusion of nuclear quantum effects. We apply a deep potential range correction (DPRc) for combined quantum mechanical/molecular mechanical (QM/MM) simulations of reactions in the condensed phase. The method uses the second-order density-functional tight-binding method (DFTB2) as a fast, approximate base QM model. The DPRc model modifies the DFTB2 QM interactions and applies short-range corrections to the QM/MM interactions to reproduce ab initio DFT (PBE0/6-31G*) QM/MM energies and forces. The DPRc thus enables both QM and QM/MM interactions to be tuned to high accuracy, and the QM/MM corrections are designed to smoothly vanish at a specified cutoff boundary (6 Å in the present work). The computational speed-up afforded by the QM/MM+DPRc model enables free energy profiles to be calculated that include rigorous long-range QM/MM interactions under periodic boundary conditions and nuclear quantum effects through a path integral approach using a new interface between the AMBER and i-PI software. The approach is demonstrated through the calculation of free energy profiles of a native RNA cleavage model reaction and reactions involving thio-substitutions, which are important experimental probes of the mechanism. The DFTB2+DPRc QM/MM free energy surfaces agree very closely with the PBE0/6-31G* QM/MM results, and it is vastly superior to the DFTB2 QM/MM surfaces with and without weighted thermodynamic perturbation corrections. 18O and 34S primary kinetic isotope effects are compared, and the influence of nuclear quantum effects on the free energy profiles is examined. Read More View Full Article Download PDF |
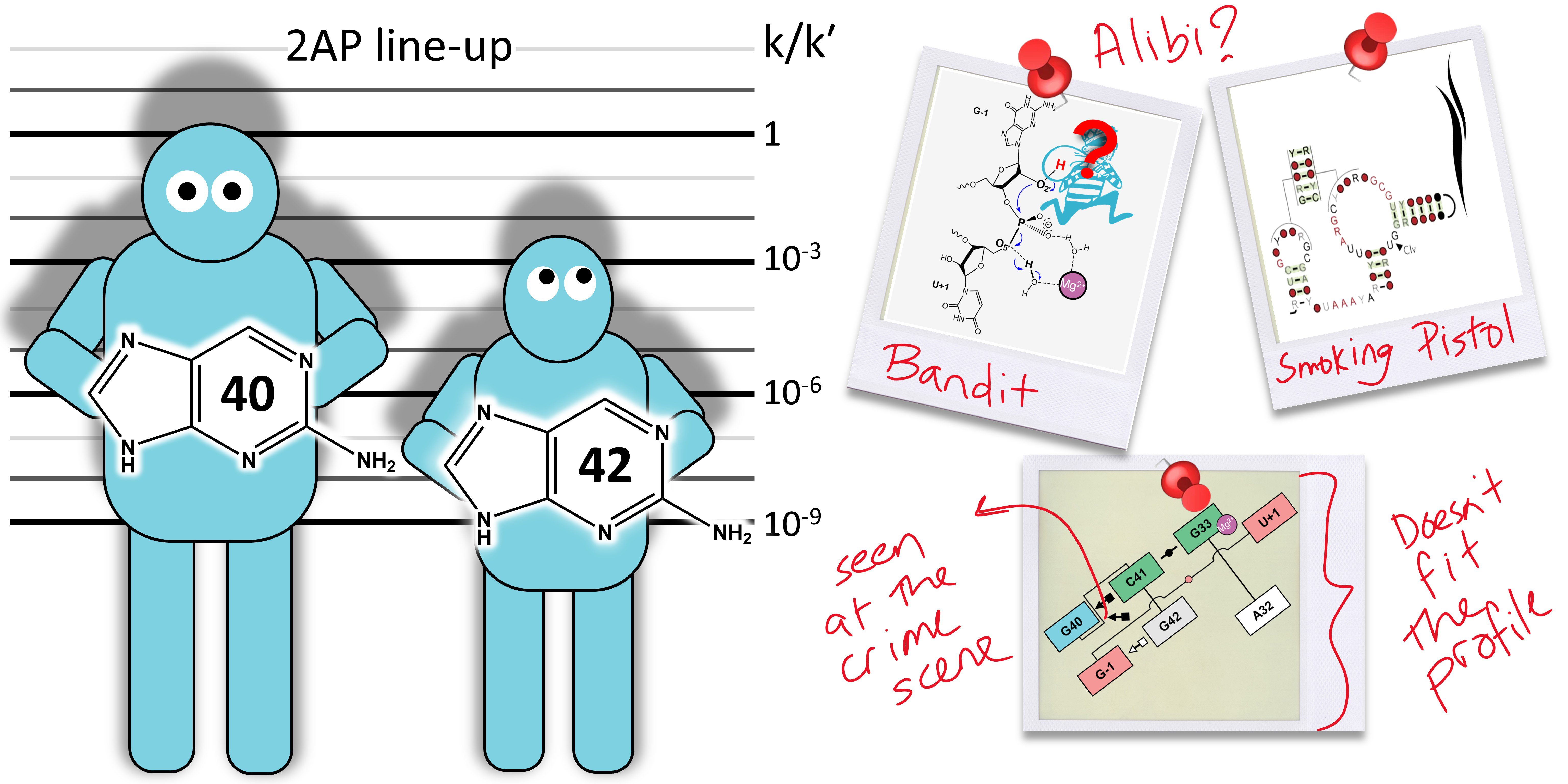 | Who stole the proton? Suspect general base guanine found with a smoking gun in the pistol ribozyme Organic & Biomolecular Chemistry (2022) 20, 6216-6230 DOI: 10.1039/d2ob00234e The pistol ribozyme (Psr) is one among the most recently discovered classes of small nucleolytic ribozymes that catalyze site-specific RNA self-cleavage through 2′-O-transphosphorylation. The Psr contains a conserved guanine (G40) that in crystal structures is in a position suggesting it plays the role of the general base to abstract a proton from the nucleophile to activate the reaction. Although some functional data is consistent with this mechanistic role, a notable exception is 2-aminopurine (2AP) substitution which has no effect on the rate, unlike similar substitutions across other so-called “G + M” and “G + A” ribozyme classes. Herein we postulate that an alternate conserved guanine, G42, is the primary general base, and provide evidence from molecular simulations that the active site of Psr can undergo local refolding into a structure that is consistent with the common “L-platform/L-scaffold” architecture identified in G + M and G + A ribozyme classes with Psr currently the notable exception. We summarize the key currently available experimental data and present new classical and combined quantum mechanical/molecular mechanical simulation results that collectively suggest a new hypothesis. We hypothesize that there are two available catalytic pathways supported by different conformational states connected by a local refolding of the active site: (1) a primary pathway with an active site architecture aligned with the L-platform/L-scaffold framework where G42 acts as a general base, and (2) a secondary pathway with the crystallographic active site architecture where G40 acts as a general base. We go on to make several experimentally testable predictions, and suggest specific experiments that would ultimately bring closure to the mystery as to “who stole the proton in the pistol ribozyme?”. Read More View Full Article Download PDF |
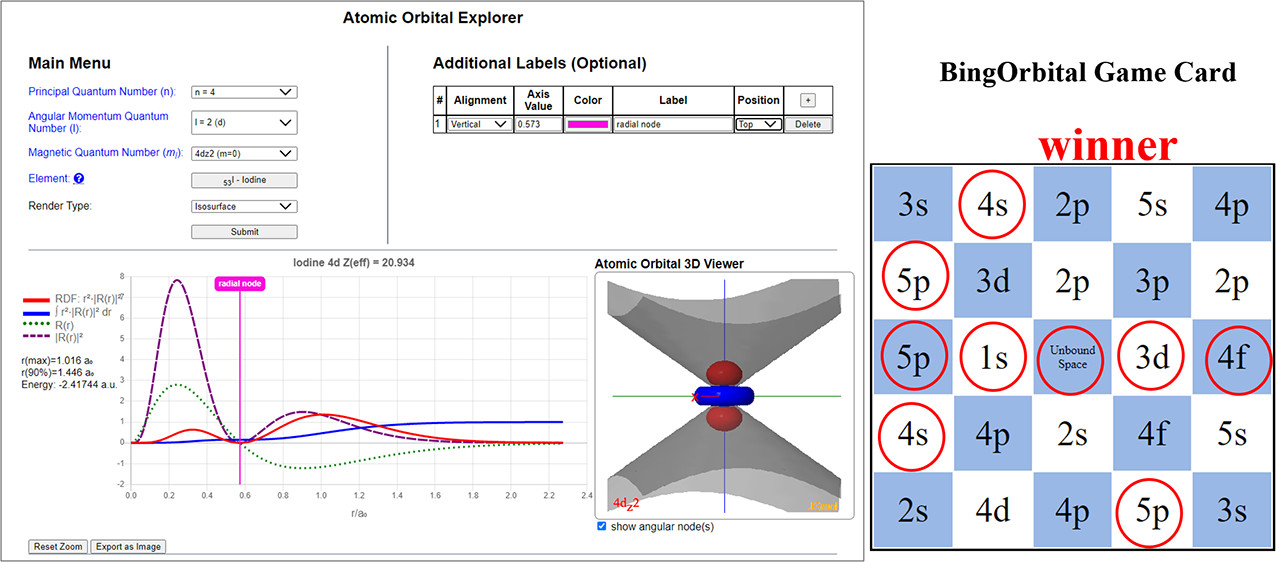 | Online Orbital Explorer and BingOrbital Game for Inquiry-Based Activities Journal of Chemical Education (2022) 99, 2135-2142 DOI: 10.1021/acs.jchemed.1c01277 We report a new online suite of tools that enables inquiry-based active-learning activities to develop students’ representational competence about atomic orbitals. Orbital Explorer is a Web site for the visualization and interactive investigation of atomic orbital properties. Orbital Explorer contains two integrated tools, namely, Atomic Orbital Explorer, which enables one to visualize and interrogate individual atomic orbitals, and Orbital RDF Comparison, which enables one to make a more detailed quantitative comparison of orbital energies and properties of orbital radial distribution functions (RDFs). In addition, we present an original chemistry educational gamification design, BingOrbital, constructed in a format resembling Bingo (American version). The game aims to reinforce the recognition of atomic orbitals based on the RDF and three-dimensional isosurface and has been applied as an engaging retrieval practice tool. A companion set of example activities that use the Orbital Explorer and BingOrbital game have been presented in another article. Read More View Full Article Download PDF |
 | Inquiry-Based Activities and Games That Engage Students in Learning Atomic Orbitals Journal of Chemical Education (2022) 99, 2175-2181 DOI: 10.1021/acs.jchemed.1c01023 Atomic orbitals represent an essential construct used to develop chemical bonding models, upon which other more advanced chemistry topics are built. In this article, we share a series of active-learning activities and a gamified approach to develop students’ representational competence about atomic orbitals and to engage students in learning the properties of atomic orbitals. These properties are essential for understanding an array of fundamental concepts such as penetration and shielding, relationships such as periodic trends, and models used to describe chemical bonding. The activities employ an inquiry-based approach to engage students in exploring the relationship between atomic orbitals’ spatial properties and quantum numbers. The activities guide students to collect data to verify periodic trends and construct electronic configurations. The activities utilize Orbital Explorer Web site for visualization, comparison, and analysis of atomic orbitals. The Orbital Explorer Web site is described in a related Technology Report. The activities and the game are suitable to be conducted in both in-person and remote-teaching settings. Read More View Full Article Download PDF |
 | Free Energy Methods in Drug Discovery—Introduction In Free Energy Methods in Drug Discovery: Current State and Future Directions (2021) 1397, 1-38 Chapter: 1 Edited by: Kira Armacost and David Thompson Publishers: ACS Publications DOI: 10.1021/bk-2021-1397.ch001 ISBN: 9780841298057 Complete understanding of most, if not all chemical processes requires at its very core the knowledge of the underlying free-energy change. In computer-aided drug design, for instance, such processes as binding of a drug to a protein or its spontaneous partitioning across the cell membrane cannot be predicted reliably without considering how the associated free energy varies. Owing to relentless theoretical developments, which have benefited from ever-growing computational resources, free-energy calculations leaning on statistical-mechanics simulations are now part of the arsenal of robust and well-characterized modeling tools. However, as will be explained below and touched upon throughout the chapters of this book, it is still challenging to obtain accurate and reliable free-energy predictions for biomolecules due to the many nuances in the system setup and the unknown unknowns such as whether a given simulation is globally converged or only locally converged, perhaps in an incorrect free-energy basin. In all but the simplest cases, free-energy simulations still require experts in the field to prepare the system, run the calculations, and analyze the results in order to obtain robust predictions that can be confidently used to make decisions in drug discovery campaigns. Read More View Full Article Download PDF |
 | Robust, Efficient and Automated Methods for Accurate Prediction of Protein-Ligand Binding Affinities in AMBER Drug Discovery Boost In Free Energy Methods in Drug Discovery: Current State and Future Directions (2021) 1397, 161-204 Chapter: 7 Edited by: Kira Armacost and David Thompson Publishers: ACS Publications DOI: 10.1021/bk-2021-1397.ch007 ISBN: 9780841298057 Recent concurrent advances in methodology development, computer hardware and simulation software has transformed our ability to make practical, quantitative predictions of relative ligand binding affinities to guide rational drug design. In the past, these calculations have been hampered by the lack of affordable software with highly efficient implementations of state-of-the-art methods on specialized hardware such as graphical processing units, combined with the paucity of available workflows to streamline throughput for real-world industry applications. Herein we discuss recent methodology development, GPU-accelerated implementation, and workflow creation for alchemical free energy simulation methods in the AMBER Drug Discovery Boost (AMBER-DD Boost) package available as a patch to AMBER20. Among the methodological advances are 1) new methods for the treatment of softcore potentials that overcome long standing end-point catastrophe and softcore imbalance problems and enable single-step alchemical transformations between ligands, 2) new adaptive enhanced sampling methods in the ”alchemical” (or ” λ”) dimension to accelerate convergence and obtain high precision ligand binding affinity predictions, 3) robust network-wide analysis methods that include cycle closure and reference constraints and restraints, and 4) practical workflows that enable streamlined calculations on large datasets to be performed. Benchmark calculations on various systems demonstrate that these tools deliver an outstanding combination of accuracy and performance, resulting in reliable high-throughput binding affinity predictions at affordable cost. Read More View Full Article |
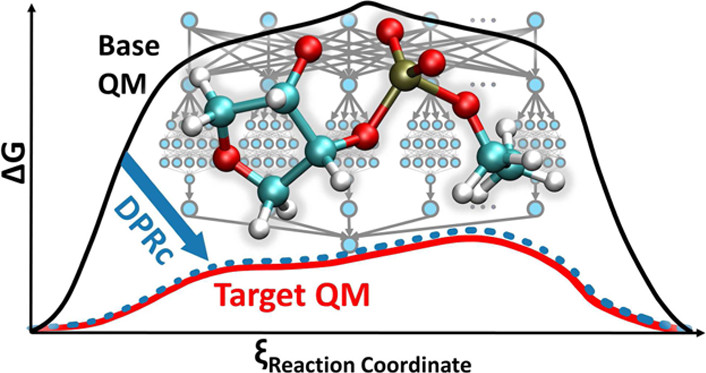 | Development of Range-Corrected Deep Learning Potentials for Fast, Accurate Quantum Mechanical/molecular Mechanical Simulations of Chemical Reactions in Solution Journal of Chemical Theory and Computation (2021) 17, 6993-7009 DOI: 10.1021/acs.jctc.1c00201 We develop a new Deep Potential - Range Correction (DPRc) machine learning potential for combined quantum mechanical/molecular mechanical (QM/MM) simulations of chemical reactions in the condensed phase. The new range correction enables short-ranged QM/MM interactions to be tuned for higher accuracy, and the correction smoothly vanishes within a specified cutoff. We further develop an active learning procedure for robust neural network training. We test the DPRc model and training procedure against a series of 6 non-enzymatic phosphoryl transfer reactions in solution that are important in mechanistic studies of RNA-cleaving enzymes. Specifically, we apply DPRc corrections to a base QM model and test its ability to reproduce free energy profiles generated from a target QM model. We perform comparisons using the MNDO/d and DFTB2 semiempirical models because they produce free energy profiles which differ significantly from each other, thereby providing us a rigorous stress test for the DPRc model and training procedure. The comparisons show that accurate reproduction of the free energy profiles requires correction of the QM/MM interactions out to 6 Å. We further find that the model's initial training benefits from generating data from temperature replica exchange simulations and including high-temperature configurations into the fitting procedure so the resulting models are trained to properly avoid high-energy regions. A single DPRc model was trained to reproduce 4 different reactions and yielded good agreement with the free energy profiles made from the target QM/MM simulations. The DPRc model was further demonstrated to be transferable to 2D free energy surfaces and 1D free energy profiles that were not explicitly considered in the training. Examination of the computational performance of the DPRc model showed that it was fairly slow when run on CPUs, but was sped up almost 100-fold when using an NVIDIA V100 GPUs, resulting in almost negligible overhead. The new DPRc model and training procedure provide a potentially powerful new tool for the creation of next-generation QM/MM potentials for a wide spectrum of free energy applications ranging from drug discovery to enzyme design. Read More View Full Article Download PDF |
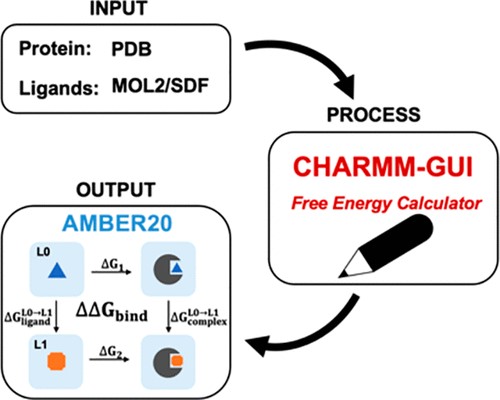 | CHARMM-GUI Free Energy Calculator for Practical Ligand Binding Free Energy Simulations with AMBER Journal of Chemical Information and Modeling (2021) 61, 4145-4151 DOI: 10.1021/acs.jcim.1c00747 Alchemical free energy methods, such as free energy perturbation (FEP) and thermodynamic integration (TI), become increasingly popular and crucial for drug design and discovery. However, the system preparation of alchemical free energy simulation is an error-prone, time-consuming, and tedious process for a large number of ligands. To address this issue, we have recently presented CHARMM-GUI Free Energy Calculator that can provide input and postprocessing scripts for NAMD and GENESIS FEP molecular dynamics systems. In this work, we extended three submodules of Free Energy Calculator to work with the full suite of GPU-accelerated alchemical free energy methods and tools in AMBER, including input and postprocessing scripts. The BACE1 (β-secretase 1) benchmark set was used to validate the AMBER-TI simulation systems and scripts generated by Free Energy Calculator. The overall results of relatively large and diverse systems are almost equivalent with different protocols (unified and split) and with different timesteps (1, 2, and 4 fs), with R2 > 0.9. More importantly, the average free energy differences between two protocols are small and reliable with four independent runs, with a mean unsigned error (MUE) below 0.4 kcal/mol. Running at least four independent runs for each pair with AMBER20 (and FF19SB/GAFF2.1/OPC force fields), we obtained a MUE of 0.99 kcal/mol and root-mean-square error of 1.31 kcal/mol for 58 alchemical transformations in comparison with experimental data. In addition, a set of ligands for T4-lysozyme was used to further validate our free energy calculation protocol whose results are close to experimental data (within 1 kcal/mol). In summary, Free Energy Calculator provides a user-friendly web-based tool to generate the AMBER-TI system and input files for high-throughput binding free energy calculations with access to the full set of GPU-accelerated alchemical free energy, enhanced sampling, and analysis methods in AMBER. Read More View Full Article Download PDF |
 | Extension of the Variational Free Energy Profile and Multistate Bennett Acceptance Ratio Methods for High-Dimensional Potential of Mean Force Profile Analysis The Journal of Physical Chemistry A (2021) 125, 4216-4232 DOI: 10.1021/acs.jpca.1c00736 We redevelop the variational free energy profile (vFEP) method using a cardinal B-spline basis to extend the method for analyzing free energy surfaces (FESs) involving three or more reaction coordinates. We also implemented software for evaluating high-dimensional profiles based on the multistate Bennett acceptance ratio (MBAR) method which constructs an unbiased probability density from global reweighting of the observed samples. The MBAR method takes advantage of a fast algorithm for solving the unbinned weighted histogram (UWHAM)/MBAR equations which replaces the solution of simultaneous equations with a nonlinear optimization of a convex function. We make use of cardinal B-splines and multiquadric radial basis functions to obtain smooth, differentiable MBAR profiles in arbitrary high dimensions. The cardinal B-spline vFEP and MBAR methods are compared using three example systems that examine 1D, 2D, and 3D profiles. Both methods are found to be useful and produce nearly indistinguishable results. The vFEP method is found to be 150 times faster than MBAR when applied to periodic 2D profiles, but the MBAR method is 4.5 times faster than vFEP when evaluating unbounded 3D profiles. In agreement with previous comparisons, we find the vFEP method produces superior FESs when the overlap between umbrella window simulations decreases. Finally, the associative reaction mechanism of hammerhead ribozyme is characterized using 3D, 4D, and 6D profiles, and the higher-dimensional profiles are found to have smaller reaction barriers by as much as 1.5 kcal/mol. The methods presented here have been implemented into the FE-ToolKit software package along with new methods for network-wide free energy analysis in drug discovery. Read More View Full Article Download PDF |
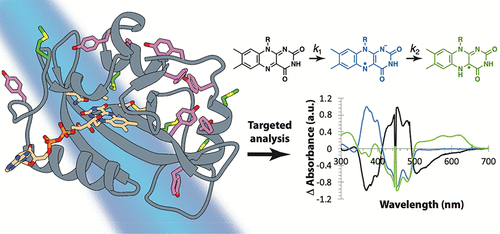 | Peripheral Methionine Residues Impact Flavin Photoreduction and Protonation in an Engineered LOV Domain Light Sensor Biochemistry (2021) 60, 1148-1164 DOI: 10.1021/acs.biochem.1c00064 Proton-coupled electron transfer reactions play critical roles in many aspects of sensory phototransduction. In the case of flavoprotein light sensors, reductive quenching of flavin excited states initiates chemical and conformational changes that ultimately transmit light signals to downstream targets. These reactions generally require neighboring aromatic residues and proton-donating side chains for rapid and coordinated electron and proton transfer to flavin. Although photoreduction of flavoproteins can produce either the anionic (ASQ) or neutral semiquinone (NSQ), the factors that favor one over the other are not well understood. Here we employ a biologically active variant of the light-oxygen-voltage (LOV) domain protein VVD devoid of the adduct-forming Cys residue (VVD-III) to probe the mechanism of flavin photoreduction and protonation. A series of isosteric and conservative residue replacements studied by rate measurements, fluorescence quantum yields, FTIR difference spectroscopy, and molecular dynamics simulations indicate that tyrosine residues facilitate charge recombination reactions that limit sustained flavin reduction, whereas methionine residues facilitate radical propagation and quenching and also gate solvent access for flavin protonation. Replacement of a single surface Met residue with Leu favors formation of the ASQ over the NSQ and desensitizes photoreduction to oxidants. In contrast, increasing site hydrophilicity by Gln substitution promotes rapid NSQ formation and weakens the influence of the redox environment. Overall, the photoreactivity of VVD-III can be understood in terms of redundant electron donors, internal hole quenching, and coupled proton transfer reactions that all depend upon protein conformation, dynamics, and solvent penetration. Read More View Full Article Download PDF |
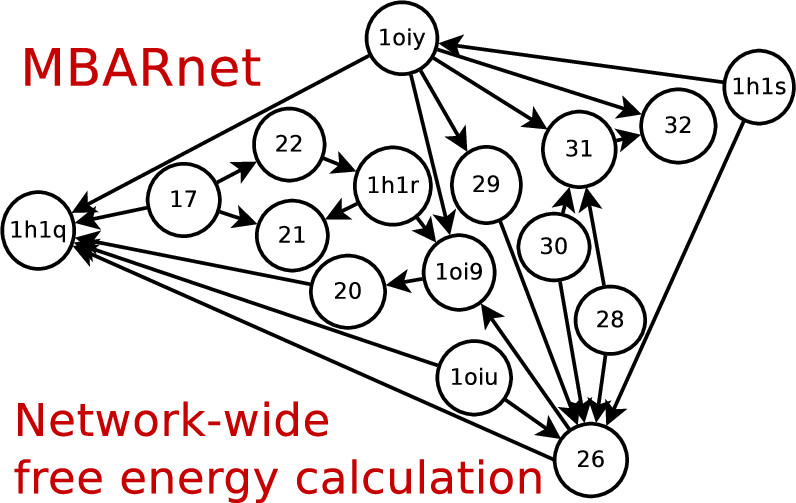 | Variational Method for Networkwide Analysis of Relative Ligand Binding Free Energies with Loop Closure and Experimental Constraints Journal of Chemical Theory and Computation (2021) 17, 1326-1336 DOI: 10.1021/acs.jctc.0c01219 We describe an efficient method for the simultaneous solution of all free energies within a relative binding free-energy (RBFE) network with cycle closure and experimental/reference constraint conditions using Bennett Acceptance Ratio (BAR) and Multistate BAR (MBAR) analysis. Rather than solving the BAR or MBAR equations for each transformation independently, the simultaneous solution of all transformations are obtained by performing a constrained minimization of a global objective function. The nonlinear optimization of the objective function is subjected to affine linear constraints that couple the free energies between the network edges. The constraints are used to enforce the closure of thermodynamic cycles within the RBFE network, and to enforce an additional set of linear constraint conditions demonstrated here to be subsets of (1 or 2) experimental values. We describe details of the practical implementation of the network BAR/MBAR procedure, including use of generalized coordinates in the minimization of the free-energy objective function, propagation of bootstrap errors from those coordinates, and performance and memory optimization. In some cases it is found that use of restraints in the optimization is more practical than use of generalized coordinates for enforcing constraint conditions. The fast BARnet and MBARnet methods are used to analyze the RBFEs of six prototypical protein–ligand systems, and it is shown that enforcement of cycle closure conditions reduces the error in the predictions only modestly, and further reduction in errors can be achieved when one or two experimental RBFEs are included in the optimization procedure. These methods have been implemented into FE-ToolKit, a new free-energy analysis toolkit. The BARnet/MBARnet framework presented here opens the door to new, more efficient and robust free-energy analysis with enhanced predictive capability for drug discovery applications. Read More View Full Article Download PDF |
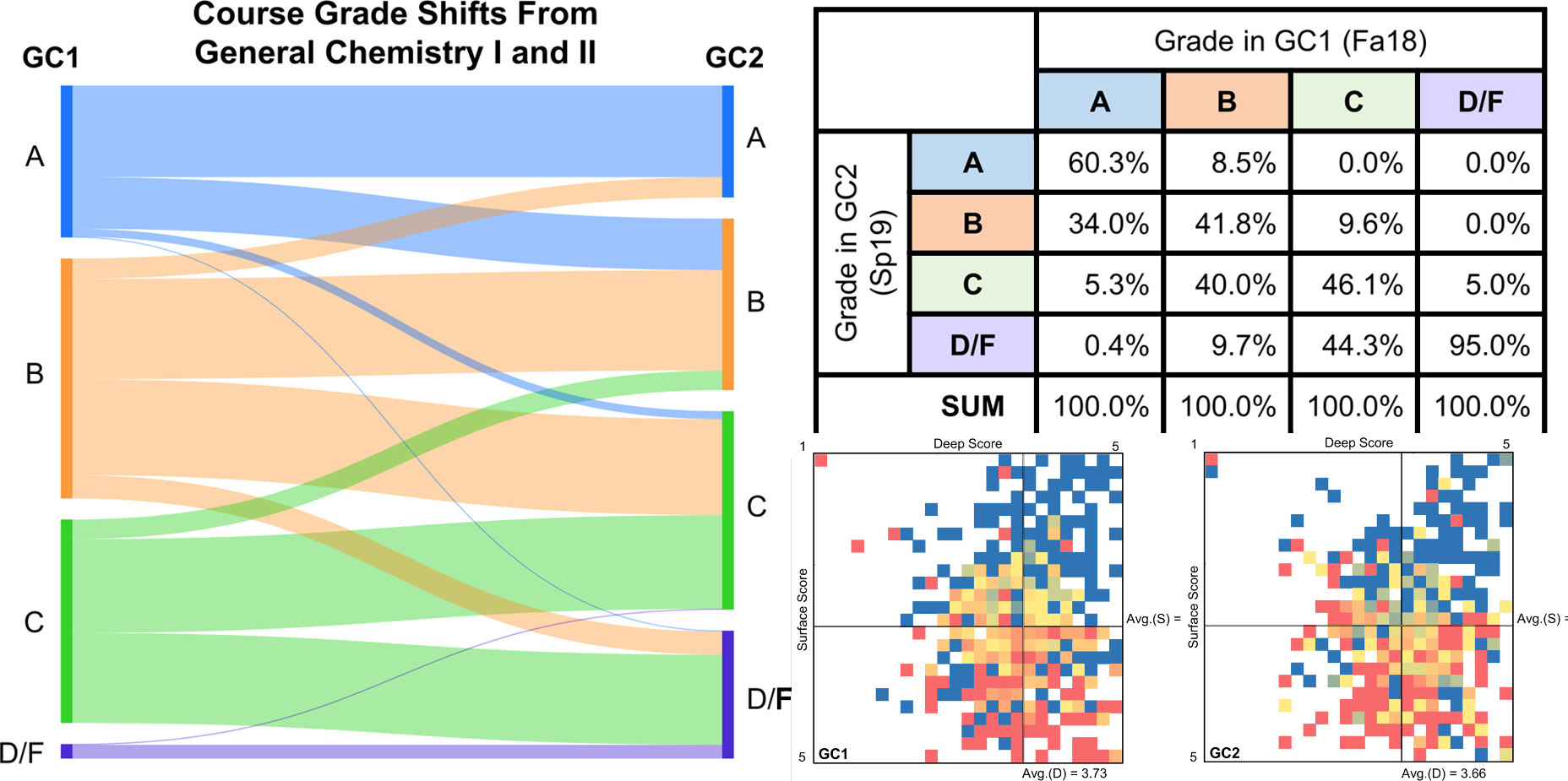 | Beneath the Surface: An Investigation of General Chemistry Students’ Study Skills to Predict Course Outcomes Journal of Chemical Education (2021) 98, 281-292 DOI: 10.1021/acs.jchemed.0c01074 As the conversation in higher education shifts from diversity to inclusion, the attrition rates of students in the STEM fields continue to be a point of discussion. Combined with the demand for expansion in the STEM workforce, various retention reforms have been proposed, implemented, and in some cases integrated into policy following evidence of success. Still, new findings, technological advances, and socio-cultural shifts inevitably necessitate an ongoing investigation as to how students approach learning. Among other factors, students who enter college without effective study skills are at much greater risk of being unsuccessful in their coursework. In order to construct an equitable learning environment, a mechanism must be developed to provide underprepared students with access to resources or interventions designed to refine the skills they need to be successful in the course. Early, reliable assessments can provide predictions of individual student outcomes in order to guide the development and implementation of such targeted interventions. In the present study, a model is developed to predict students’ odds of success based on their study approaches, as measured by their responses to twelve survey items from an existing instrument used in the Chemistry Education Research literature designed to measure students’ deep and surface learning approaches. The model’s prediction specificity ranges from 66.5% to 86.9% by semester. Two distinct sets of lower-performing students are identified in the data: those who align predominantly with surface approaches to learning versus those who indicate using both deep and surface approaches to learning. This supports the idea of a tailored approach to interventions, rather than a one-size-fits-all solution. Results from this instrument were correlated to students’ reported study methods and beliefs. Read More View Full Article Download PDF |
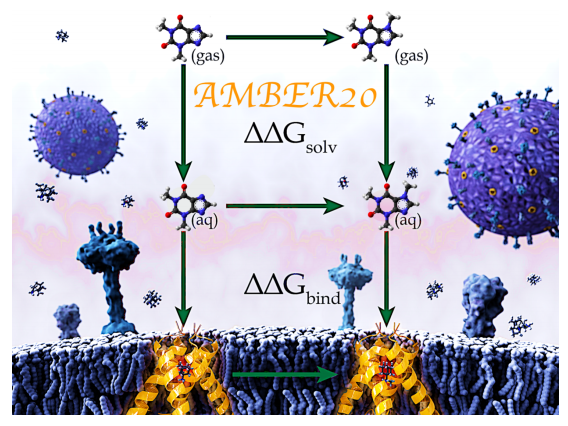 | Alchemical Binding Free Energy Calculations in AMBER20: Advances and Best Practices for Drug Discovery Journal of Chemical Information and Modeling (2020) 60, 5595-5623 DOI: 10.1021/acs.jcim.0c00613 Predicting protein-ligand binding affinities and the associated thermodynamics of biomolecular recognition is a primary objective of structure-based drug design. Alchemical free energy simulations offer a highly accurate and computationally efficient route to achieving this goal. While the AMBER molecular dynamics package has successfully been used for alchemical free energy simulations in academic research groups for decades, widespread impact in industrial drug discovery settings has been minimal due to previous limitations within the AMBER alchemical code, coupled with challenges in system setup and postprocessing workflows. Through a close academia-industry collaboration we have addressed many of the previous limitations with an aim to improve accuracy, efficiency and robustness of alchemical binding free energy simulations in industrial drug discovery applications. Here, we highlight some of the recent advances in AMBER20 with a focus on alchemical binding free energy (BFE) calculations, which are less computationally intensive than alternative binding free energy methods where full binding/unbinding paths are explored. In addition to scientific and technical advances in AMBER20, we also describe the essential practical aspects associated with running relative alchemical BFE calculations along with recommendations for best practices, highlighting the importance not only of the alchemical simulation code, but also the auxiliary functionalities and expertise required to obtain accurate and reliable results. This work is intended to provide a contemporary overview of the scientific, technical, and practical issues associated with running relative BFE simulations in AMBER20, with a focus on real-world drug discovery applications. Read More View Full Article Download PDF |
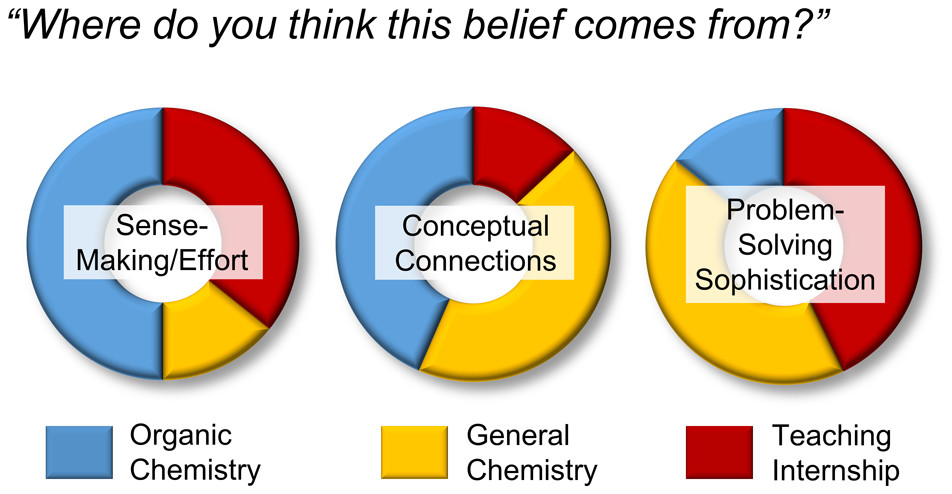 | Through the Looking CLASS: When Peer Leader Learning Attitudes Are Not What They Seem Journal of Chemical Education (2020) 97, 2078-2090 DOI: 10.1021/acs.jchemed.0c00129 The Teaching Internship is a credit-bearing program composed of undergraduate near peer instructors (teaching interns, or TIs) that offers supplemental assistance for students in the General Chemistry courses. With fellow undergraduates serving as a role model and student–faculty liaison, the benefits of near peer instruction have been well-documented. Because TIs develop a dual role of student and instructor over time, they afford a unique opportunity to explore the middle area of the expert/novice spectrum. Identifying the most influential components of the TI role may allow practitioners to implement these components in other ways for different groups of students. The present work provides a description of the TI model and uses a mixed-methods approach to analyze how the peer leadership role impacted the TIs’ attitudes about learning chemistry. Quantitative results show that TIs do hold predominantly expert-like learning attitudes compared to the General Chemistry population from which they are selected; however, evidence of novice thinking is still observed in some areas. This survey data was then used to inform a qualitative approach. Further analysis indicated that TIs’ responses on survey items were context-dependent, and that peer leadership experiences were associated with expert learning attitudes and appear to be influential in the development of these attitudes. These findings suggest that these factors should be taken into account when drawing general conclusions from survey results. Read More View Full Article Download PDF |
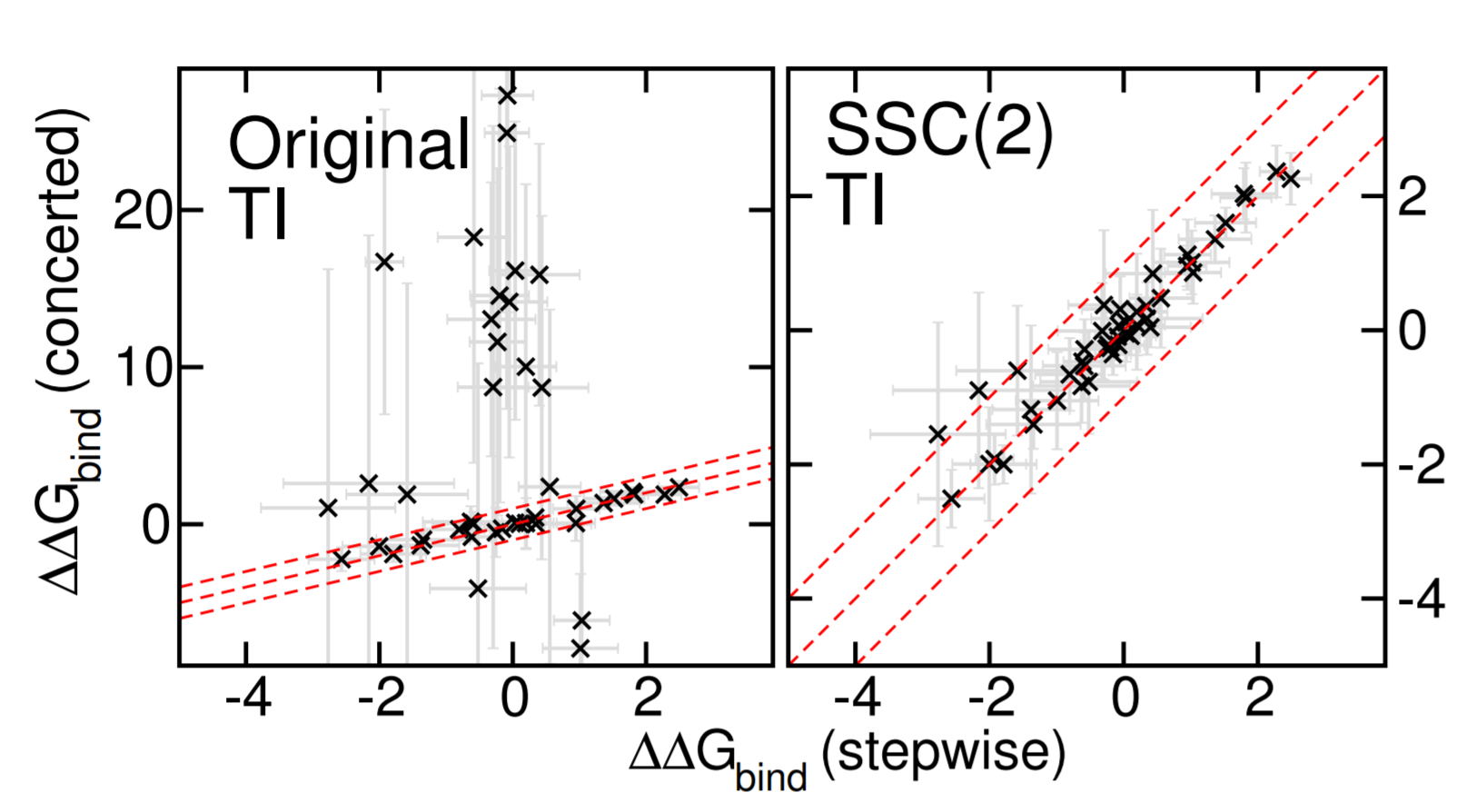 | Improved Alchemical Free Energy Calculations with Optimized Smoothstep Softcore Potentials Journal of Chemical Theory and Computation (2020) 16, 5512-5525 DOI: 10.1021/acs.jctc.0c00237 Progress in the development of GPU-accelerated free energy simulation software has enabled practical applications on complex biological systems and fueled efforts to develop more accurate and robust predictive methods. In particular, this work re-examines concerted (a.k.a., one-step or unified) alchemical transformations commonly used in the prediction of hydration and relative binding free energies (RBFEs). We first classify several known challenges in these calculations into three categories: endpoint catastrophes, particle collapse, and large gradient-jumps. While endpoint catastrophes have long been addressed using softcore potentials, the remaining two problems occur much more sporadically and can result in either numerical instability (i.e. complete failure of a simulation) or inconsistent estimation (i.e. stochastic convergence to an incorrect result). The particle collapse problem stems from an imbalance in short-range electrostatic and repulsive interactions and can, in principle, be solved by appropriately balancing the respective softcore parameters. However, the large gradient-jump problem itself arises from the sensitivity of the free energy to large values of the softcore parameters, as might be used in trying to solve the particle collapse issue. Often no satisfactory compromise exists with the existing softcore potential form. As a framework for solving these problems, we developed a new family of smoothstep softcore (SSC) potentials motivated by an analysis of the derivatives along the alchemical path. The smoothstep polynomials generalize the monomial functions that are used in most implementations and provide an additional path-dependent smoothing parameter. The effectiveness of this approach is demonstrated on simple, yet pathological cases that illustrate the three problems outlined. With appropriate parameter selection we find that a second-order SSC(2) potential does at least as well as the conventional approach and provides a vast improvement in terms of consistency across all cases. Lastly, we compare the concerted SSC(2) approach against the gold-standard stepwise (a.k.a., decoupled or multi-step) scheme over a large set of RBFE calculations as might be encountered in drug discovery. Read More View Full Article Download PDF |
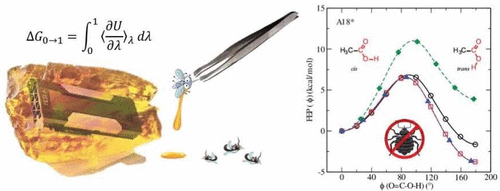 | Validation of Free Energy Methods in AMBER Journal of Chemical Information and Modeling (2020) 60, 5296-5300 DOI: 10.1021/acs.jcim.0c00285 With advancements in GPU-accelerated free energy methods, it is now possible to obtain sufficiently high precision in free energy calculations to rigorously stress test implementations for consistency, reproducibility and reliability. Herein we rovide high precision validation tests that examine alchemical transformations of a small molecule data set that has been used elsewhere to examine the reproducibility of free energy calculations across different molecular simulation software packages. We demonstrate that the most recent, updated AMBER18 provides consistent free energy results in both the gas phase and in solution. We first show, in the context of thermodynamic integration (TI), that results are invariant with respect to “split” (e.g., stepwise decharge-vdW-recharge) versus “unified” protocols. This brought to light a subtle inconsistency in previous versions of AMBER that was traced to the improper treatment of 1-4 vdW and electrostatic interactions involving atoms across the softcore boundary. We illustrate that, under the assumption that the ensembles produced by different legs of the alchemical transformation between molecules “A” and “B” in the gas phase and aqueous phase are very small, the inconsistency on the relative hydration free energy is minimal. However, for general cases where the ensembles are shown to be substantially different, these errors can be large. Finally, we demonstrate that results for relative hydration free energy simulations are independent of TI or multistate Bennett’s acceptance ratio (MBAR) analysis, invariant to the specific choice of the softcore region, and agree with results derived from absolute hydration free energy values. Read More View Full Article Download PDF |
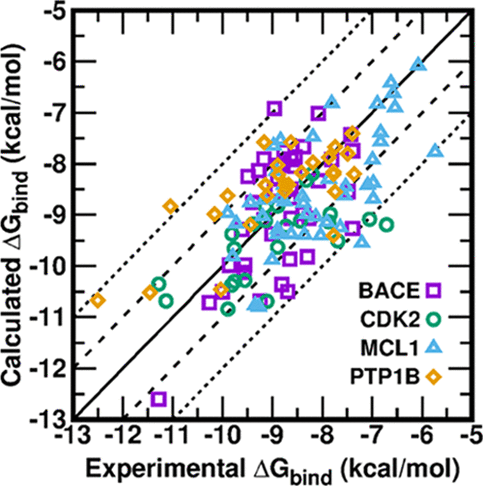 | Fast, Accurate, and Reliable Protocols for Routine Calculations of Protein−Ligand Binding Affinities in Drug Design Projects Using AMBER GPU-TI with ff14SB/GAFF ACS Omega (2020) 5, 4611-4619 DOI: 10.1021/acsomega.9b04233 Accurate prediction of the absolute or relative protein−ligand binding affinity is one of the major tasks in computer-aided drug design projects, especially in the stage of lead optimization. In principle, the alchemical free energy (AFE) methods such as thermodynamic integration (TI) or free-energy perturbation (FEP) can fulfill this task, but in practice, a lot of hurdles prevent them from being routinely applied in daily drug design projects, such as the demanding computing resources, slow computing processes, unavailable or inaccurate force field parameters, and difficult and unfriendly setting up and post-analysis procedures. In this study, we have exploited practical protocols of applying the CPU (central processing unit)-TI and newly developed GPU (graphic processing unit)-TI modules and other tools in the AMBER software package, combined with ff14SB/GAFF1.8 force fields, to conduct efficient and accurate AFE calculations on protein−ligand binding free energies. We have tested 134 protein−ligand complexes in total for four target proteins (BACE, CDK2, MCL1, and PTP1B) and obtained overall comparable performance with the commercial Schrodinger FEP+ program (Wang et al. J. Am. Chem. Soc. 2015, 137, 2695−2703). The achieved accuracy fits within the requirements for computations to generate effective guidance for experimental work in drug lead optimization, and the needed wall time is short enough for practical application. Our verified protocol provides a practical solution for routine AFE calculations in real drug design projects. Read More View Full Article Download PDF |
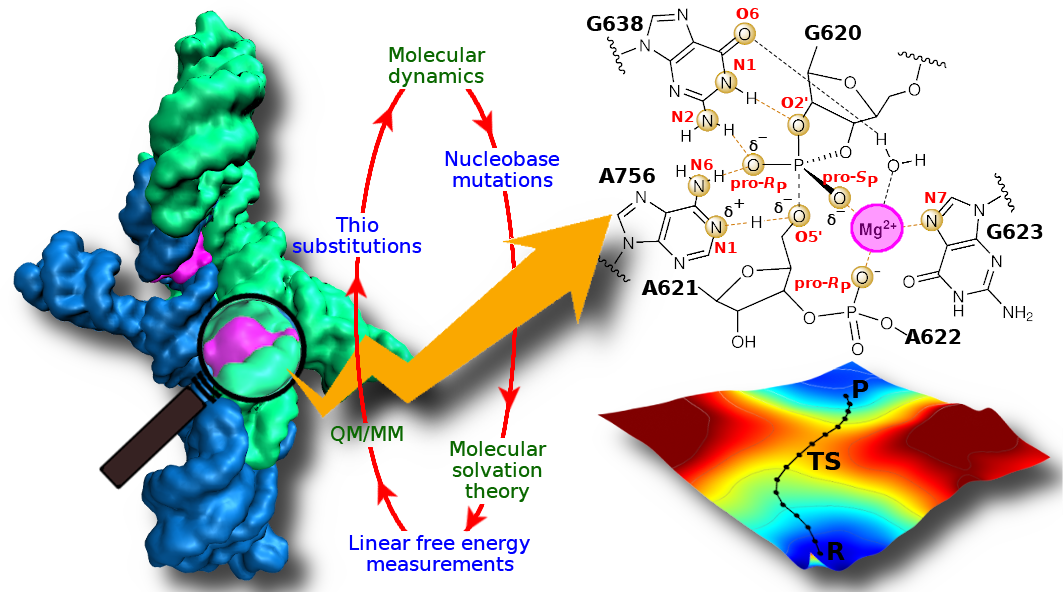 | Confluence of theory and experiment reveals the catalytic mechanism of the Varkud satellite ribozyme Nature Chemistry (2020) 12, 193-201 DOI: 10.1038/s41557-019-0391-x The Varkud satellite ribozyme catalyses site-specific RNA cleavage and ligation, and serves as an important model system to understand RNA catalysis. Here, we combine stereospecific phosphorothioate substitution, precision nucleobase mutation and linear free-energy relationship measurements with molecular dynamics, molecular solvation theory and ab initio quantum mechanical/molecular mechanical free-energy simulations to gain insight into the catalysis. Through this confluence of theory and experiment, we unify the existing body of structural and functional data to unveil the catalytic mechanism in unprecedented detail, including the degree of proton transfer in the transition state. Further, we provide evidence for a critical Mg2+ in the active site that interacts with the scissile phosphate and anchors the general base guanine in position for nucleophile activation. This novel role for Mg2+ adds to the diversity of known catalytic RNA strategies and unifies functional features observed in the Varkud satellite, hairpin and hammerhead ribozyme classes. Read More View Full Article Download PDF |
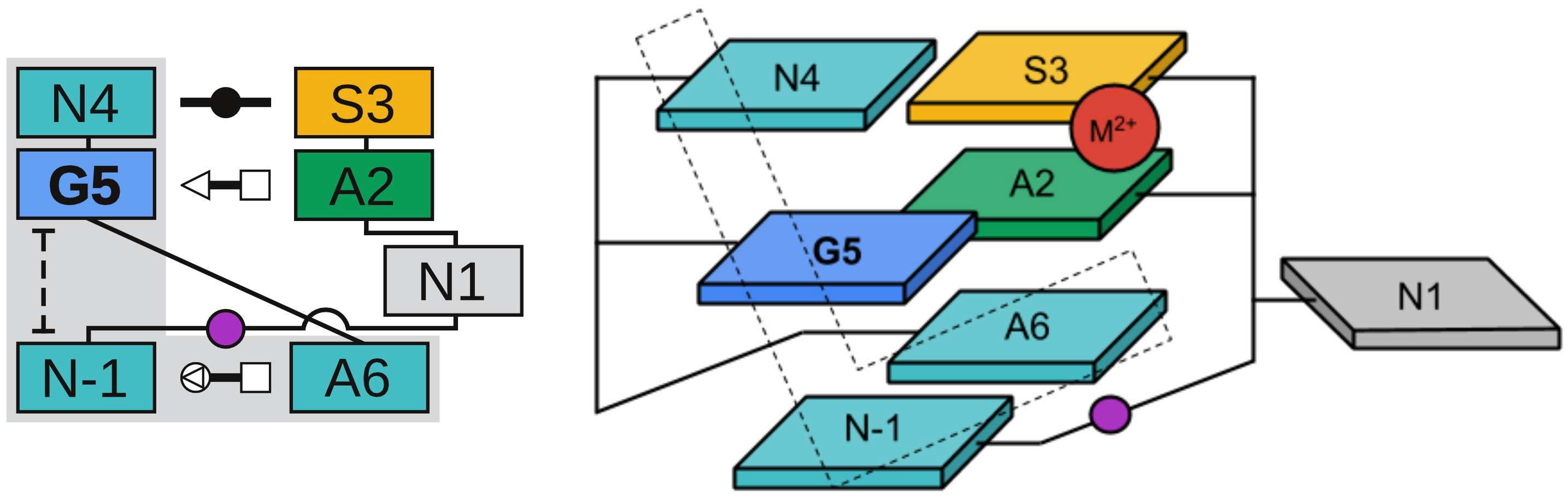 | The L-platform/L-scaffold framework: a blueprint for RNA-cleaving nucleic acid enzyme design RNA (2020) 26, 111-126 DOI: 10.1261/rna.071894.119 We develop an L-platform/L-scaffold framework we hypothesize may serve as a blueprint to facilitate site-specific RNA-cleaving nucleic acid enzyme design. Building on the L-platform motif originally described by Suslov and coworkers, we identify new critical scaffolding elements required to anchor a conserved general base guanine ("L-anchor") and bind functionally important metal ions at the active site ("L-pocket"). Molecular simulations, together with a broad range of experimental structural and functional data, connect the L-platform/L-scaffold elements to necessary and sufficient conditions for catalytic activity. We demonstrate that the L-platform/L-scaffold framework is common to 5 of the 9 currently known naturally occurring ribozyme classes (Twr, HPr, VSr, HHr, Psr), and intriguingly from a design perspective, the framework also appears in an artificially engineered DNAzyme (8-17dz). The flexibility of the L-platform/L-scaffold framework is illustrated on these systems, highlighting modularity and trends in the variety of known general acid moieties that are supported. These trends give rise to two distinct catalytic paradigms, building on the classifications proposed by Wilson and coworkers and named for the implicated general base and acid. The "G+A" paradigm (Twr, HPr, VSr) exclusively utilizes nucleobase residues for chemistry, and the "G+M+" paradigm (HHr, 8-17dz, Psr) involves structuring of the "L-pocket" metal ion binding site for recruitment of a divalent metal ion that plays an active role in the chemical steps of the reaction. Finally, the modularity of the L-platform/L-scaffold framework is illustrated in the VS ribozyme where the "L-pocket" assumes the functional role of the "L-anchor" element, highlighting a distinct mechanism, but one that is functionally linked with the hammerhead ribozyme. Read More View Full Article Download PDF |
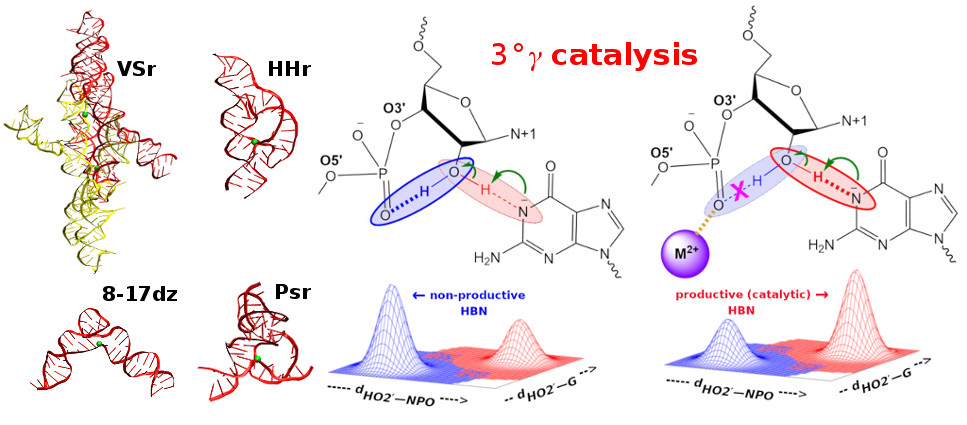 | Evidence for a Catalytic Strategy to Promote Nucleophile Activation in Metal-Dependent RNA-Cleaving Ribozymes and 8-17 DNAzyme ACS Catalysis (2019) 9, 10612-10617 DOI: 10.1021/acscatal.9b02035 An unique catalytic strategy was recently reported for the glmS ribozyme [Bingaman et al., Nat. Chem. Biol. 2017, 13, 439−445] that involves promotion of productive hydrogen bonding of the O2′ nucleophile to facilitate its activation. We provide broad evidence of this strategy in the hammerhead, pistol, and VS ribozymes and 8-17 DNAzyme, enabled by a functionally important divalent metal ion that interacts with the scissile phosphate and disrupts nonproductive competitive hydrogen bonding with the O2′ nucleophile. This strategy, designated tertiary gamma (3°γ) catalysis, illustrates an additional role for divalent ions in ribozyme catalysis. Read More View Full Article Download PDF |
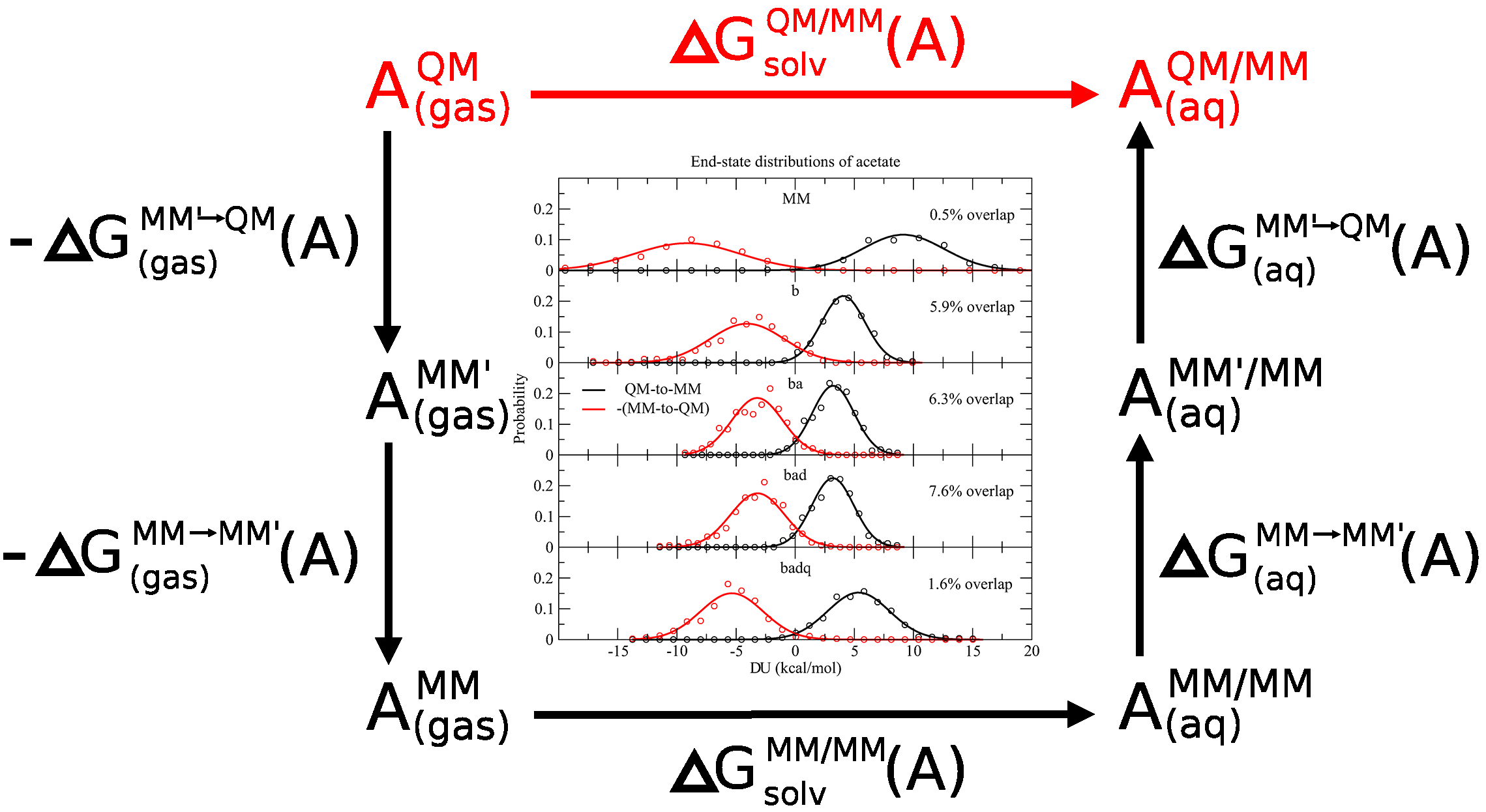 | Development of a Robust Indirect Approach for MM → QM Free Energy Calculations That Combines Force-Matched Reference Potential and Bennett’s Acceptance Ratio Methods Journal of Chemical Theory and Computation (2019) 15, 5543-5562 DOI: 10.1021/acs.jctc.9b00401 We use the PBE0/6-31G* density functional method to perform ab initio quantum mechanical/molecular mechanical (QM/MM) molecular dynamics (MD) simulations under periodic boundary conditions with rigorous electrostatics using the ambient potential composite Ewald method in order to test the convergence of MM → QM/MM free energy corrections for the prediction of 17 small-molecule solvation free energies and eight ligand binding free energies to T4 lysozyme. The “indirect” thermodynamic cycle for calculating free energies is used to explore whether a series of reference potentials improve the statistical quality of the predictions. Specifically, we construct a series of reference potentials that optimize a molecular mechanical (MM) force field’s parameters to reproduce the ab initio QM/MM forces from a QM/MM simulation. The optimizations form a systematic progression of successively expanded parameters that include bond, angle, dihedral, and charge parameters. For each reference potential, we calculate benchmark quality reference values for the MM → QM/MM correction by performing the mixed MM and QM/MM Hamiltonians at 11 intermediate states, each for 200 ps. We then compare forward and reverse application of Zwanzig’s relation, thermodynamic integration (TI), and Bennett’s acceptance ratio (BAR) methods as a function of reference potential, simulation time, and the number of simulated intermediate states. We find that Zwanzig’s equation is inadequate unless a large number of intermediate states are explicitly simulated. The TI and BAR mean signed errors are very small even when only the end-state simulations are considered, and the standard deviations of the TI and BAR errors are decreased by choosing a reference potential that optimizes the bond and angle parameters. We find a robust approach for the data sets of fairly rigid molecules considered here is to use bond + angle reference potential together with the end-state-only BAR analysis. This requires QM/MM simulations to be performed in order to generate reference data to parametrize the bond + angle reference potential, and then this same simulation serves a dual purpose as the full QM/MM end state. The convergence of the results with respect to time suggests that computational resources may be used more efficiently by running multiple simulations for no more than 50 ps, rather than running one long simulation. Read More View Full Article Download PDF |
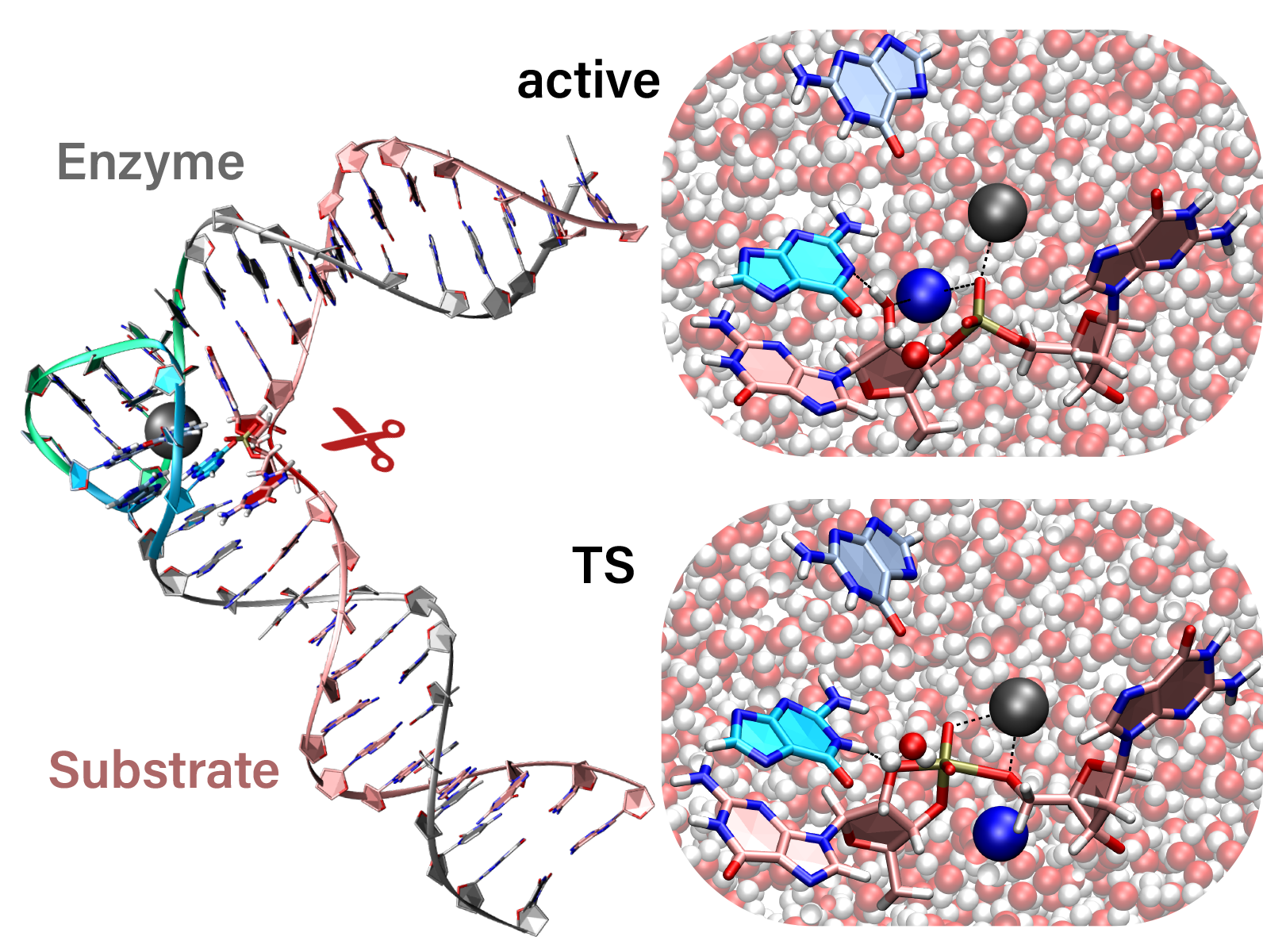 | Dynamical ensemble of the active state and transition state mimic for the RNA-cleaving 8–17 DNAzyme in solution Nucleic Acids Research (2019) 47, 10282-10295 DOI: 10.1093/nar/gkz773 We perform molecular dynamics simulations, based on recent crystallographic data, on the 8–17 DNAzyme at four states along the reaction pathway to determine the dynamical ensemble for the active state and transition state mimic in solution. A striking finding is the diverse roles played by Na+ and Pb2+ ions in the electrostatically strained active site that impact all four fundamental catalytic strategies, and share commonality with some features recently inferred for naturally occurring hammerhead and pistol ribozymes. The active site Pb2+ ion helps to stabilize in-line nucleophilic attack, provides direct electrostatic transition state stabilization, and facilitates leaving group departure. A conserved guanine residue is positioned to act as the general base, and is assisted by a bridging Na+ ion that tunes the pKa and facilitates in-line fitness. The present work provides insight into how DNA molecules are able to solve the RNA-cleavage problem, and establishes functional relationships between the mechanism of these engineered DNA enzymes with their naturally evolved RNA counterparts. This adds valuable information to our growing body of knowledge on general mechanisms of phosphoryl transfer reactions catalyzed by RNA, proteins and DNA. Read More View Full Article Download PDF |
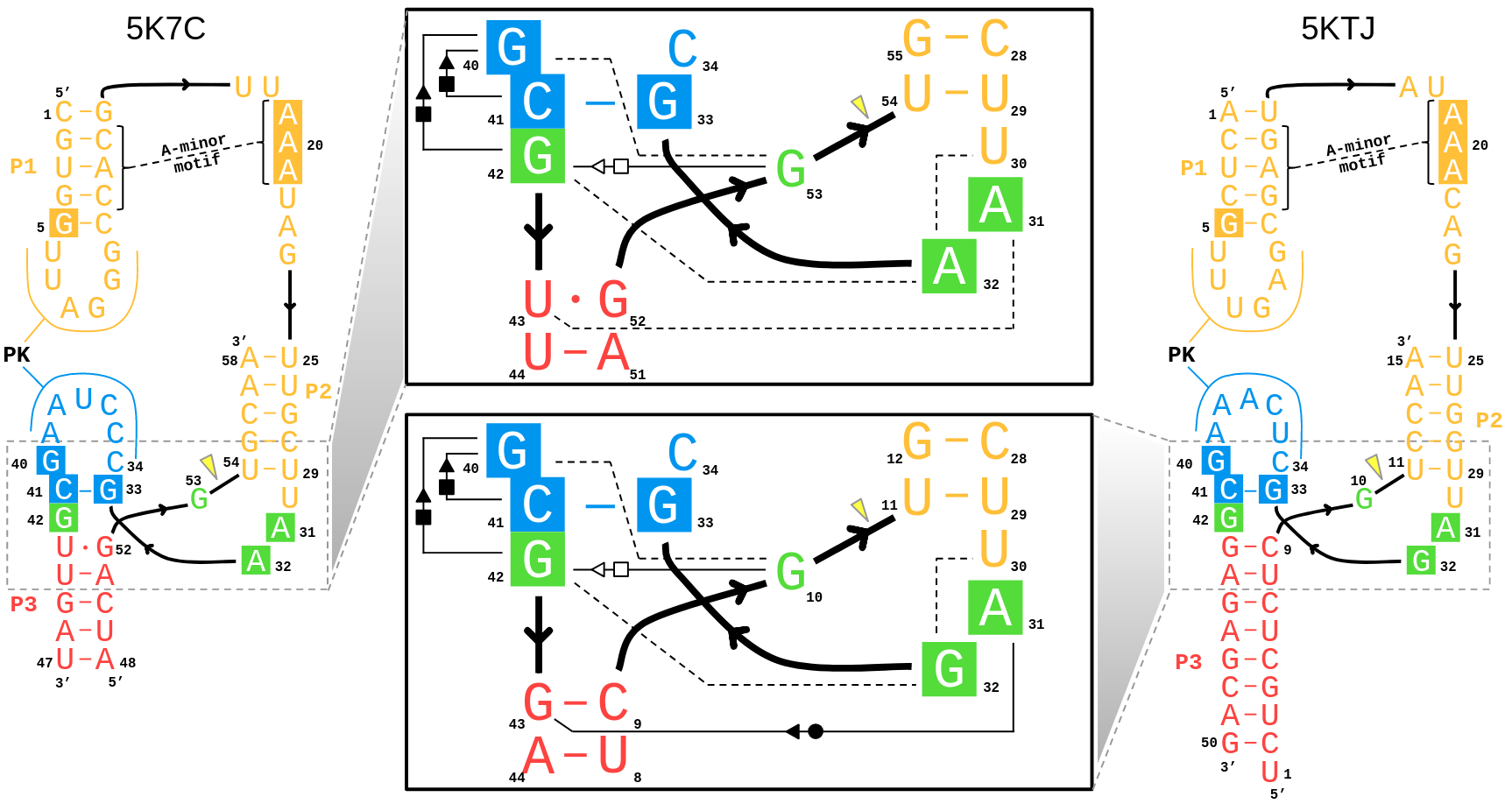 | Molecular simulations of the pistol ribozyme: unifying the interpretation of experimental data and establishing functional links with the hammerhead ribozyme RNA (2019) 25, 1439-1456 DOI: 10.1261/rna.071944.119 The pistol ribozyme (Psr) is among the most recently discovered RNA enzymes, and has been the subject of experiments aimed at elucidating mechanism. Recent biochemical studies have revealed exciting clues about catalytic interactions in the active site not apparent from available crystallographic data. The present work unifies the interpretation of the existing body of structural and functional data on Psr by providing a dynamical model for the catalytically active state in solution from molecular simulation. Our results suggest that a catalytic Mg2+ ion makes inner-sphere contact with G33:N7 and outer-sphere coordination to the pro-Rp of the scissile phosphate, promoting electrostatic stabilization of the dianionic transition state and neutralization of the developing charge of the leaving group through a metal-coordinated water molecule that is made more acidic by a hydrogen bond donated from the 2’OH of P32. This model is consistent with experimental activity-pH and mutagenesis data, including sensitivity to G33(7cG) and phosphorothioate substitution/metal ion rescue. The model suggests several experimentally testable predictions including stereospecific thio substitutions and G42X (X=xanthine) mutations, some of which have appeared and been validated during the review of this work. Further, the model identifies striking similarities of Psr to the hammerhead ribozyme (HHr), including: similar global fold, organization of secondary structure around an active site 3-way junction, catalytic metal ion binding mode, and guanine general base. However, the specific binding mode and role of the Mg2+ ion, as well as a conserved 2’-OH in the active site, are inter-related but subtly different between the ribozymes. Read More View Full Article Download PDF |
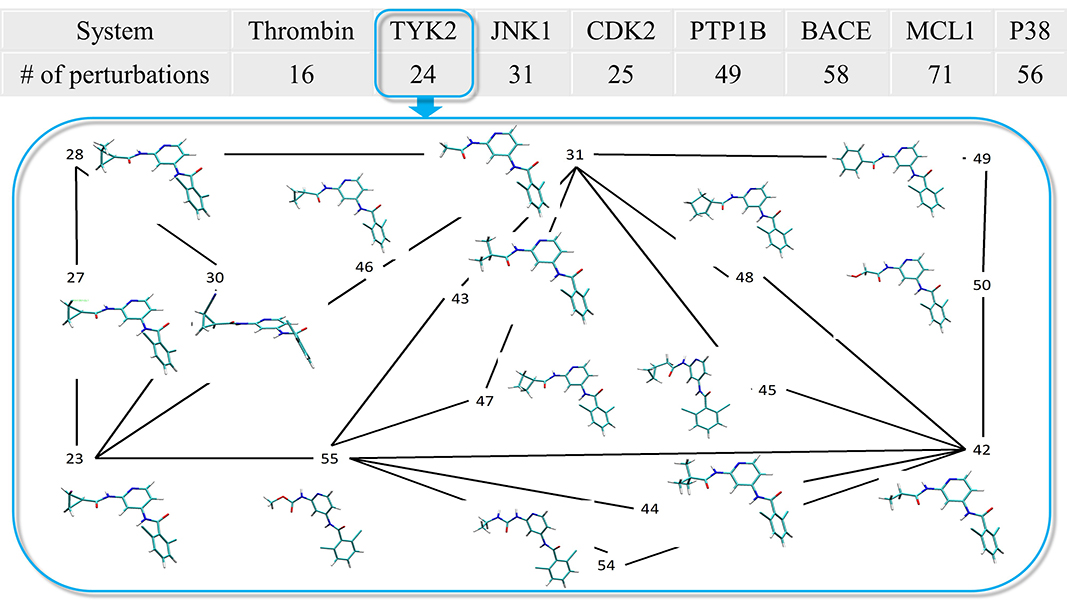 | Using AMBER18 for Relative Free Energy Calculations Journal of Chemical Information and Modeling (2019) 59, 3128-3135 DOI: 10.1021/acs.jcim.9b00105 With renewed interest in free energy methods in contemporary structure-based drug design, there is a pressing need to validate against multiple targets and force fields to assess the overall ability of these methods to accurately predict relative binding free energies. We computed relative binding free energies using graphics processing unit accelerated thermodynamic integration (GPU-TI) on a data set originally assembled by Schrödinger, Inc. Using their GPU free energy code (FEP+) and the OPLS2.1 force field combined with the REST2 enhanced sampling approach, these authors obtained an overall MUE of 0.9 kcal/mol and an overall RMSD of 1.14 kcal/mol. In our study using GPU-TI from AMBER with the AMBER14SB/GAFF1.8 force field but without enhanced sampling, we obtained an overall MUE of 1.17 kcal/mol and an overall RMSD of 1.50 kcal/mol for the 330 perturbations contained in this data set. A more detailed analysis of our results suggested that the observed differences between the two studies arise from differences in sampling protocols along with differences in the force fields employed. Future work should address the problem of establishing benchmark quality results with robust statistical error bars obtained through multiple independent runs and enhanced sampling, which is possible with the GPU-accelerated features in AMBER. Read More View Full Article Download PDF |
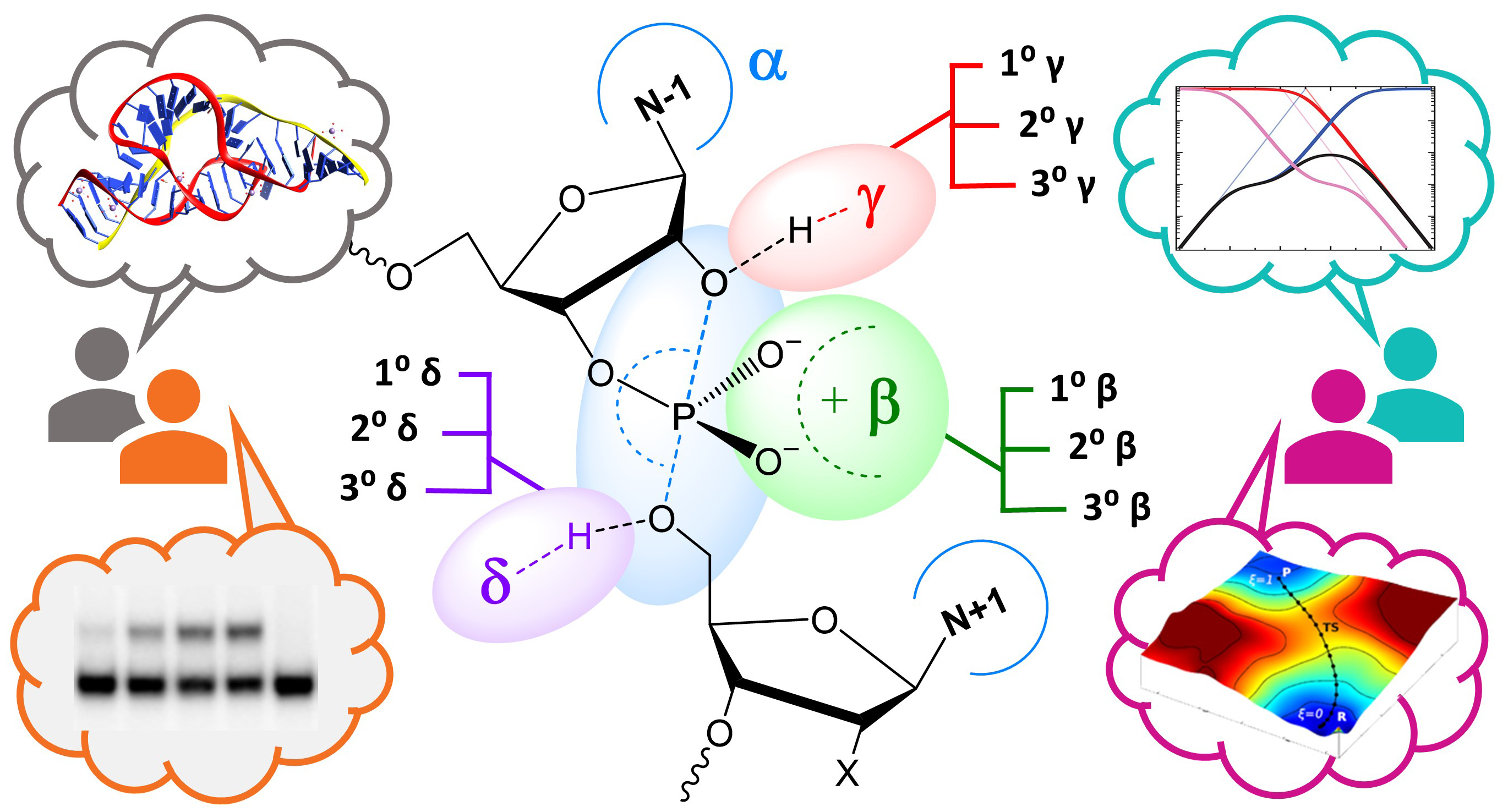 | An Ontology for Facilitating Discussion of Catalytic Strategies of RNA-Cleaving Enzymes ACS Chemical Biology (2019) 14, 1068-1076 DOI: 10.1021/acschembio.9b00202 PMC6661149 A predictive understanding of the mechanisms of RNA cleavage is important for the design of emerging technology built from biological and synthetic molecules that have promise for new biochemical and medicinal applications. Over the past 15 years, RNA cleavage reactions involving 2′-O-transphosphorylation have been discussed using a simplified framework introduced by Breaker that consists of four fundamental catalytic strategies (designated α, β, γ, and δ) that contribute to rate enhancement. As more detailed mechanistic data emerge, there is need for the framework to evolve and keep pace. We develop an ontology for discussion of strategies of enzymes that catalyze RNA cleavage via 2′-O-transphosphorylation that stratifies Breaker’s framework into primary (1°), secondary (2°), and tertiary (3°) contributions to enable more precise interpretation of mechanism in the context of structure and bonding. Further, we point out instances where atomic-level changes give rise to changes in more than one catalytic contribution, a phenomenon we refer to as “functional blurring”. We hope that this ontology will help clarify our conversations and pave the path forward toward a consensus view of these fundamental and fascinating mechanisms. The insight gained will deepen our understanding of RNA cleavage reactions catalyzed by natural protein and RNA enzymes, as well as aid in the design of new engineered DNA and synthetic enzymes. Read More View Full Article Download PDF |
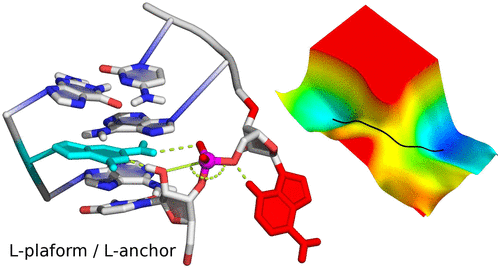 | Cleaning Up Mechanistic Debris Generated by Twister Ribozymes Using Computational RNA Enzymology ACS Catalysis (2019) 9, 5803-5815 DOI: 10.1021/acscatal.9b01155 PMC6641568 The catalytic properties of RNA have been a subject of fascination and intense research since their discovery over 30 years ago. Very recently, several classes of nucleolytic ribozymes have emerged and been characterized structurally. Among these, the twister ribozyme has been center-stage and a topic of debate about its architecture and mechanism owing to conflicting interpretations of different crystal structures and in some cases conflicting interpretations of the same functional data. In the present work, we attempt to clean up the mechanistic "debris" generated by twister ribozymes using a comprehensive computational RNA enzymology approach aimed to provide a unified interpretation of existing structural and functional data. Simulations in the crystalline environment and in solution provide insight into the origins of observed differences in crystal structures and coalesce on a common active site architecture and dynamical ensemble in solution. We use GPU-accelerated free energy methods with enhanced sampling to ascertain microscopic nucleobase pKa values of the implicated general acid and base, from which predicted activity–pH profiles can be compared directly with experiments. Next, ab initio quantum mechanical/molecular mechanical (QM/MM) simulations with full dynamic solvation under periodic boundary conditions are used to determine mechanistic pathways through multidimensional free energy landscapes for the reaction. We then characterize the rate-controlling transition state and make predictions about kinetic isotope effects and linear free energy relations. Computational mutagenesis is performed to explain the origin of rate effects caused by chemical modifications and to make experimentally testable predictions. Finally, we provide evidence that helps to resolve conflicting issues related to the role of metal ions in catalysis. Throughout each stage, we highlight how a conserved L-platform structural motif, together with a key L-anchor residue, forms the characteristic active site scaffold enabling each of the catalytic strategies to come together for not only the twister ribozyme but also the majority of the known small nucleolytic ribozyme classes. Read More View Full Article Download PDF |
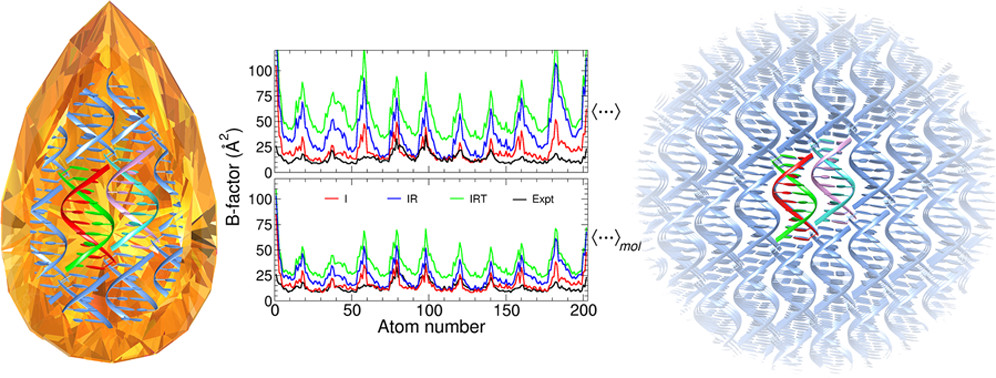 | Framework for Conducting and Analyzing Crystal Simulations of Nucleic Acids to Aid in Modern Force Field Evaluation The Journal of Physical Chemistry B (2019) 123, 4611-4624 DOI: 10.1021/acs.jpcb.8b11923 PMC6614744 Crystal simulations provide useful tools, along with solution simulations, to test nucleic acid force fields, but should be interpreted with care owing to the difficulty of establishing the environmental conditions needed to reproduce experimental crystal packing. These challenges underscore the need to construct proper protocols for carrying out crystal simulations and analyzing results to identify the origin of deviations from crystallographic data. Toward this end, we introduce a novel framework for B-factor decomposition into additive intramolecular, rotational, and translational atomic fluctuation components and partitioning of each of these components into individual asymmetric unit and lattice contributions. We apply the framework to a benchmark set of A-DNA, Z-DNA, and B-DNA double helix systems of various chain lengths. Overall, the intramolecular deviations from the crystal were quite small (≤1.0 Å), suggesting high accuracy of the force field, whereas crystal packing was not well reproduced. The present work establishes a framework to conduct and analyze crystal simulations that ultimately take on issues of crystal packing and can provide insight into nucleic acid force fields. Read More View Full Article Download PDF |
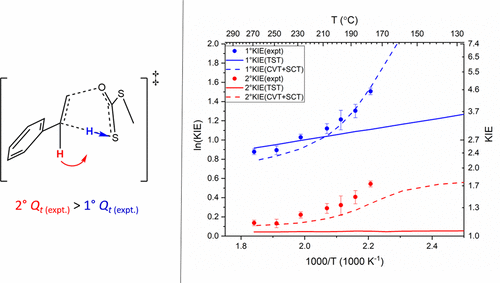 | Quantum Suppression of Intramolecular Deuterium Kinetic Isotope Effects in a Pericyclic Hydrogen Transfer Reaction The Journal of Physical Chemistry A (2019) 123, 3647-3654 DOI: 10.1021/acs.jpca.9b00172 PMC6661151 It is generally accepted that hydrogen tunneling enhances both primary and secondary H/D kinetic isotope effects (KIEs) over what would be expected under the assumptions of classical barrier transition. Previous studies have exclusively shown that the effects of tunneling upon primary H/D KIEs have been much larger than those observed for secondary H/D KIEs. Here we present a series of experimental H/D KIE results associated with the Chugaev elimination of methyl xanthate derived from ß-phenylethanol over the temperature range of 180 to 290 °C. Intramolecular H/D KIEs computed according to classical transition state theory (TST) are markedly overestimated relative to experimentally measured values. Experimental intermolecular H/D KIEs and direct dynamic calculations based on canonical variational transition state theory (CVT) with small-curvature tunneling correction (SCT) reveal that this result is largely the consequence of extraordinary tunneling enhancement of the secondary H/D KIE. This unexpected behavior is examined in the context of other similar hydrogen transfer reactions. Read More View Full Article Download PDF |
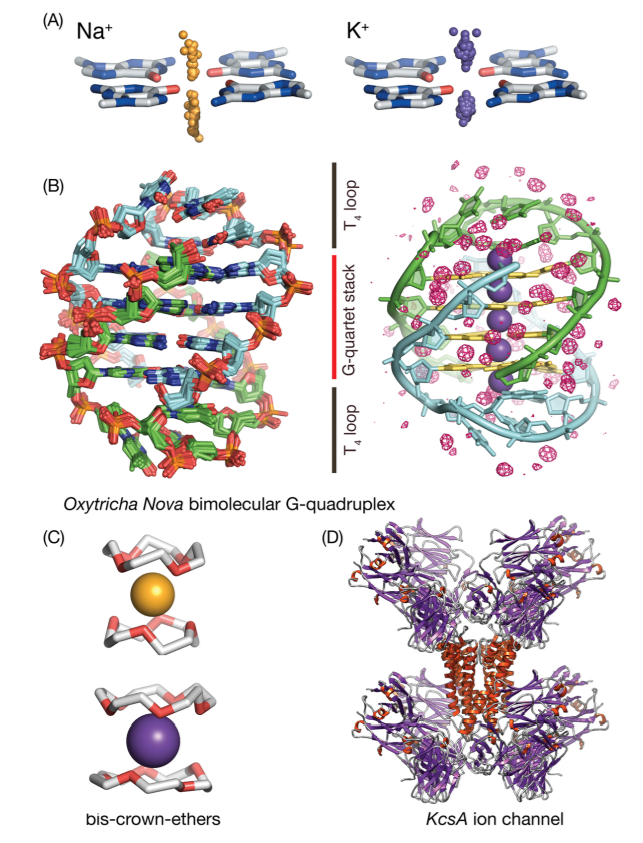 | Predicting site-binding modes of ions and water to nucleic acids using molecular solvation theory Journal of the American Chemical Society (2019) 141, 2435 DOI: 10.1021/jacs.8b11474 PMC6574206 Abstract Site binding of ions and water shapes nucleic acids folding, dynamics and biological function, complementing the more diffuse territorial binding. Unlike territorial binding, prediction of site-specific binding to nucleic acids remains an unsolved challenge in computational biophysics. This work presents a new toolset based on the 3D-RISM molecular solvation theory and topological analysis that predicts cation and water site-binding to nucleic acids. 3D-RISM is shown to accurately capture alkali cations and water binding to the central channel, transversal loops and grooves of the Oxytricha nova's telomeres G-quadruplex (Oxy-GQ), in agreement with high resolution crystallographic data. To improve the computed cations occupancy along the Oxy-GQ central channel, it was necessary to refine and validate new cation-oxygen parameters using structural and thermodynamic data available for crown ethers and ion channels. This single set of parameters that describes both localized and delocalized binding to various biological systems is used to gain insight into cation occupancy along the Oxy-GQ channel under various salt conditions. The paper concludes with prospects for extending the method to predict divalent metals binding to nucleic acids. This work advances the forefront of theoretical methods able to provide predictive insight into ion atmosphere effects on nucleic acids function. Read More View Full Article Download PDF |
 | A Comparison of QM/MM Simulations with and without the Drude Oscillator Model Based on Hydration Free Energies of Simple Solutes Molecules (2018) 23, 2695-2719 DOI: 10.3390/molecules23102695 PMC6222909 Maintaining a proper balance between specific intermolecular interactions and non-specific solvent interactions is of critical importance in molecular simulations, especially when predicting binding affinities or reaction rates in the condensed phase. The most rigorous metric for characterizing solvent affinity are solvation free energies, which correspond to a transfer from the gas phase into solution. Due to the drastic change of the electrostatic environment during this process, it is also a stringent test of polarization response in the model. Here, we employ both the CHARMM fixed charge and polarizable force fields to predict hydration free energies of twelve simple solutes. The resulting classical ensembles are then reweighted to obtain QM/MM hydration free energies using a variety of QM methods, including MP2, Hartree–Fock, density functional methods (BLYP, B3LYP, M06-2X) and semi-empirical methods (OM2 and AM1 ). Our simulations test the compatibility of quantum-mechanical methods with molecular-mechanical water models and solute Lennard–Jones parameters. In all cases, the resulting QM/MM hydration free energies were inferior to purely classical results, with the QM/MM Drude force field predictions being only marginally better than the QM/MM fixed charge results. In addition, the QM/MM results for different quantum methods are highly divergent, with almost inverted trends for polarizable and fixed charge water models. While this does not necessarily imply deficiencies in the QM models themselves, it underscores the need to develop consistent and balanced QM/MM interactions. Both the QM and the MM component of a QM/MM simulation have to match, in order to avoid artifacts due to biased solute–solvent interactions. Finally, we discuss strategies to improve the convergence and efficiency of multi-scale free energy simulations by automatically adapting the molecular-mechanics force field to the target quantum method. Read More View Full Article Download PDF |
 | GPU-Accelerated Molecular Dynamics and Free Energy Methods in Amber18: Performance Enhancements and New Features Journal of Chemical Information and Modeling (2018) 58, 2043-2050 DOI: 10.1021/acs.jcim.8b00462 PMC6226240 We report progress in graphics processing unit (GPU)-accelerated molecular dynamics and free energy methods in Amber18. Of particular interest is the development of alchemical free energy algorithms, including free energy perturbation and thermodynamic integration methods with support for nonlinear soft-core potential and parameter interpolation transformation pathways. These methods can be used in conjunction with enhanced sampling techniques such as replica exchange, constant-pH molecular dynamics, and new 12–6–4 potentials for metal ions. Additional performance enhancements have been made that enable appreciable speed-up on GPUs relative to the previous software release. Read More View Full Article Download PDF |
 | On the convergence of multi-scale free energy simulations Molecular Simulation (2018) 44, 1062-1081 DOI: 10.1080/08927022.2018.1475741 PMC6298030 In this work, we employ simple model systems to evaluate the relative performance of two of the most important free energy methods: The Zwanzig equation (also known as "Free energy perturbation") and Bennett’s acceptance ratio method (BAR). Although our examples should be transferable to other kinds of free energy simulations, we focus on applications of multi-scale free energy simulations. Such calculations are especially complex, since they connect two different levels of theory with very different requirements in terms of speed, accuracy, sampling and parallelisability. We try to reconcile all those different factors by developing some simple criteria to guide the early stages of the development of a free energy protocol. This is accomplished by quantifying how many intermediate steps and how many potential energy evaluations are necessary in order to reach a certain level of convergence. Read More View Full Article Download PDF |
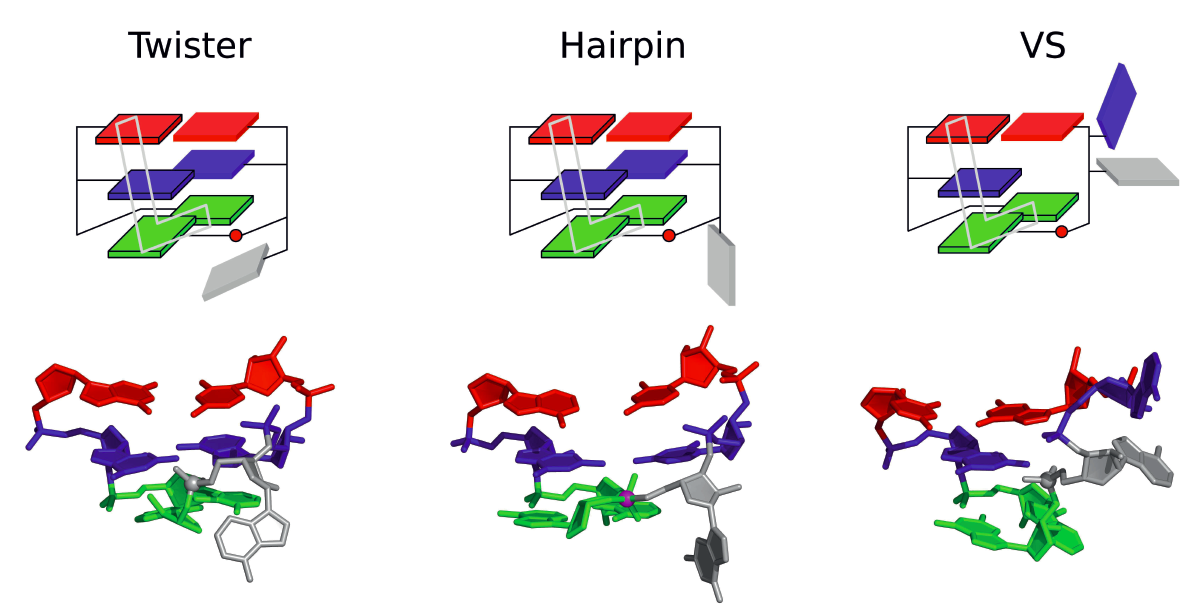 | Catalytic Strategies of Nucleolytic Ribozymes Catalysis in Chemistry and Biology (Proceedings of the 24th International Solvay Conference on Chemistry)(2018) Session 6, 341-344 DOI: 10.1142/10907 The ability of RNA molecules to selectively and efficiently catalyze complex chemical transformation has vast implications. Our understanding of the mechanisms of RNA catalysis have been greatly advanced by the study of small nucleolytic RNA enzymes, or ribozymes, that have evolved naturally in viruses and living organisms, or artificially through high-throughput in vitro selection techniques [1]. Experimental structural and mechanistic work, along with computational simulations have provided deep insight into the mechanism of these model ribozyme systems. Very recently, there has been a surge in progress in the determination of crystallographic structures of ribozymes [2–6] that have provided a departure point for theoretical investigations that aim to bridge the gap between structural and mechanistic measurements, and provide a detailed dynamical picture of mechanism at atomic-level resolution. In this way, molecular simulations have the potential to unify the interpretations of a broad range of experimental data and establish a consensus view of mechanism [7]. Ultimately, multiscale simulations, together with experiments, afford the tools needed to gain predictive insight into catalysis, including control factors that regulate selectivity and reactivity, that may guide rational design efforts. In the present work, results from multiscale molecular simulations are presented for a series of ribozymes for which crystallographic data has recently become available, including the twister [2], Varkud satallite virus (VS) [3] and pistol [4] ribozymes. These results uncover recurring themes, as well as new twists, in the catalytic strategies taken by ribozymes that are apparent only when broadly analyzing their structure, biochemical characterization and detailed mechanisms predicted by molecular simulations. The interpretations of experimental data afforded by new multiscale molecular simulation results uncover general principles and provide predictive insight into the catalytic mechanisms of nucleolytic ribozymes that may guide rational design efforts. Read More View Full Article Download PDF |
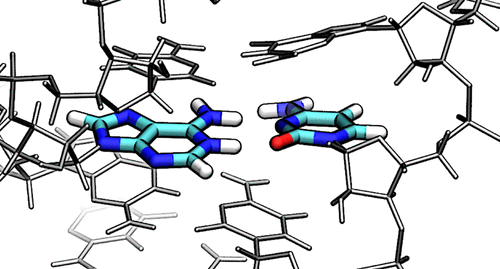 | A GPU-Accelerated Parameter Interpolation Thermodynamic Integration Free Energy Method Journal of Chemical Theory and Computation (2018) 14, 1564-1582 DOI: 10.1021/acs.jctc.7b01175 PMC5849537There has been a resurgence of interest in free energy methods motivated by the performance enhancements offered by molecular dynamics (MD) software written for specialized hardware, such as graphics processing units (GPUs). In this work, we exploit the properties of a parameter-interpolated thermodynamic integration (PI-TI) method to connect states by their molecular mechanical (MM) parameter values. This pathway is shown to be better behaved for Mg2+ ? Ca2+ transformations than traditional linear alchemical pathways (with and without soft-core potentials). The PI-TI method has the practical advantage that no modification of the MD code is required to propagate the dynamics, and unlike with linear alchemical mixing, only one electrostatic evaluation is needed (e.g., single call to particle-mesh Ewald) leading to better performance. In the case of AMBER, this enables all the performance benefits of GPU-acceleration to be realized, in addition to unlocking the full spectrum of features available within the MD software, such as Hamiltonian replica exchange (HREM). The TI derivative evaluation can be accomplished efficiently in a post-processing step by reanalyzing the statistically independent trajectory frames in parallel for high throughput. We also show how one can evaluate the particle mesh Ewald contribution to the TI derivative evaluation without needing to perform two reciprocal space calculations. We apply the PI-TI method with HREM on GPUs in AMBER to predict pKa values in double stranded RNA molecules and make comparison with experiments. Convergence to under 0.25 units for these systems required 100 ns or more of sampling per window and coupling of windows with HREM. We find that MM charges derived from ab initio QM/MM fragment calculations improve the agreement between calculation and experimental results. Read More View Full Article Download PDF |
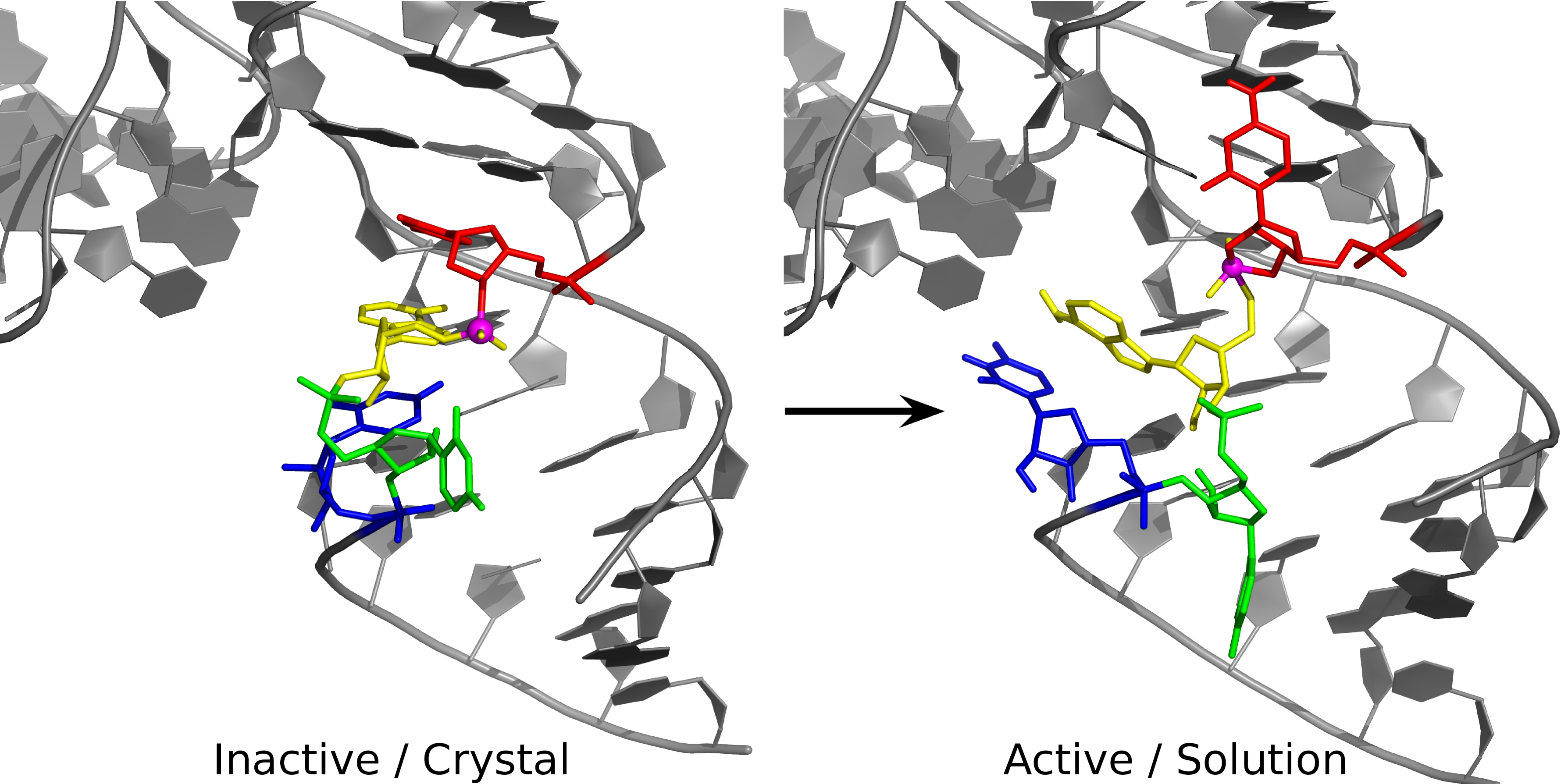 | Model for the Functional Active State of the TS Ribozyme from Molecular Simulation Angewandte Chemie International Edition (2017) 56, 13392-13395 DOI: 10.1002/anie.201705608 PMC5843185 Recently, a crystal structure has been reported of a new catalytic RNA, the TS ribozyme, that has been identified through comparative genomics and is believed to be a metalloribozyme having novel mechanistic features. Although this data provides invaluable structural information, analysis suggests a conformational change is required to arrive at a catalytically relevant state. We report results of molecular simulations that predict a spontaneous local rearrangement of the active site, leading to solution structures consistent with available functional data and providing competing mechanistic hypotheses that can be experimentally tested. The two competing hypotheses differ in the proposed identity of the catalytic general acid: either a water molecule coordinating a Mg2+ ion bound at the Watson-Crick edge of C7, or the N3 position of residue C7 itself. Read More View Full Article Download PDF |
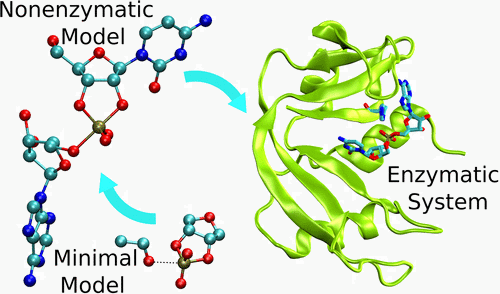 | A Multidimensional B-Spline Correction for Accurate Modeling Sugar Puckering in QM/MM Simulations Journal of Chemical Theory and Computation (2017) 13, 3975-3984 DOI: 10.1021/acs.jctc.7b00161 PMC5839098 The computational efficiency of approximate quantum mechanical methods allows their use for the construction of multidimensional reaction free energy profiles. It has recently been demonstrated that quantum models based on the neglect of diatomic differential overlap (NNDO) approximation have difficulty modeling deoxyribose and ribose sugar ring puckers and thus limit their predictive value in the study of RNA and DNA systems. A method has been introduced in our previous work to improve the description of the sugar puckering conformational landscape that uses a multidimensional B-spline correction map (BMAP correction) for systems involving intrinsically coupled torsion angles. This method greatly improved the adiabatic potential energy surface profiles of DNA and RNA sugar rings relative to high-level ab initio methods even for highly problematic NDDO-based models. In the present work, a BMAP correction is developed, implemented, and tested in molecular dynamics simulations using the AM1/d-PhoT semiempirical Hamiltonian for biological phosphoryl transfer reactions. Results are presented for gas-phase adiabatic potential energy surfaces of RNA transesterification model reactions and condensed-phase QM/MM free energy surfaces for nonenzymatic and RNase A-catalyzed transesterification reactions. The results show that the BMAP correction is stable, efficient, and leads to improvement in both the potential energy and free energy profiles for the reactions studied, as compared with ab initio and experimental reference data. Exploration of the effect of the size of the quantum mechanical region indicates the best agreement with experimental reaction barriers occurs when the full CpA dinucleotide substrate is treated quantum mechanically with the sugar pucker correction. Read More View Full Article Download PDF |
 | Quantum mechanical force fields for condensed phase molecular simulations Journal of Physics: Condensed Matter (2017) 29, 383002-383016 DOI: 10.1088/1361-648X/aa7c5c PMC5821073 Molecular simulations are powerful tools for providing atomic-level details into complex chemical and physical processes that occur in the condensed phase. For strongly interacting systems where quantum many-body effects are known to play an important role, density-functional methods are often used to provide the model with the potential energy used to drive dynamics. These methods, however, suffer from two major drawbacks. First, they are often too computationally intensive to practically apply to large systems over long time scales, limiting their scope of application. Second, there remain challenges for these models to obtain the necessary level of accuracy for weak non-bonded interactions to obtain quantitative accuracy for a wide range of condensed phase properties. Quantum mechanical force fields (QMFFs) provide a potential solution to both of these limitations. In this review, we address recent advances in the development of QMFFs for condensed phase simulations. In particular, we examine the development of QMFF models using both approximate and ab initio density-functional models, the treatment of short-ranged non-bonded and long-ranged electrostatic interactions, and stability issues in molecular dynamics calculations. Example calculations are provided for crystalline systems, liquid water, and ionic liquids. We conclude with a perspective for emerging challenges and future research directions. Read More View Full Article Download PDF |
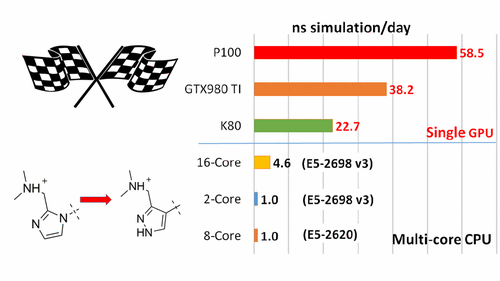 | Toward Fast and Accurate Binding Affinity Prediction with pmemdGTI: An Efficient Implementation of GPU-Accelerated Thermodynamic Integration Journal of Chemical Theory and Computation (2017) 13, 3077-3084 DOI: 10.1021/acs.jctc.7b00102 PMC5843186 We report the implementation of the thermodynamic integration method on the pmemd module of the AMBER 16 package on GPUs (pmemdGTI). The pmemdGTI code typically delivers over 2 orders of magnitude of speed-up relative to a single CPU core for the calculation of ligand-protein binding affinities with no statistically significant numerical differences and thus provides a powerful new tool for drug discovery applications. Read More View Full Article Download PDF |
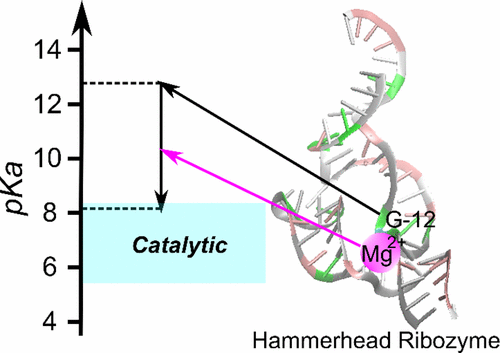 | Divalent Metal Ion Activation of a Guanine General Base in the Hammerhead Ribozyme: Insights from Molecular Simulations Biochemistry (2017) 56, 2985-2994 DOI: 10.1021/acs.biochem.6b01192 PMC5832362 The hammerhead ribozyme is a well-studied nucleolytic ribozyme that catalyzes the self-cleavage of the RNA phosphodiester backbone. Despite experimental and theoretical efforts, key questions remain about details of the mechanism with regard to the activation of the nucleophile by the putative general base guanine (G12). Straightforward interpretation of the measured activity–pH data implies the pKa value of the N1 position in the G12 nucleobase is significantly shifted by the ribozyme environment. Recent crystallographic and biochemical work has identified pH-dependent divalent metal ion binding at the N7/O6 position of G12, leading to the hypothesis that this binding mode could induce a pKa shift of G12 toward neutrality. We present computational results that support this hypothesis and provide a model that unifies the interpretation of available structural and biochemical data and paints a detailed mechanistic picture of the general base step of the reaction. Experimentally testable predictions are made for mutational and rescue effects on G12, which will give further insights into the catalytic mechanism. These results contribute to our growing knowledge of the potential roles of divalent metal ions in RNA catalysis. Read More View Full Article Download PDF |
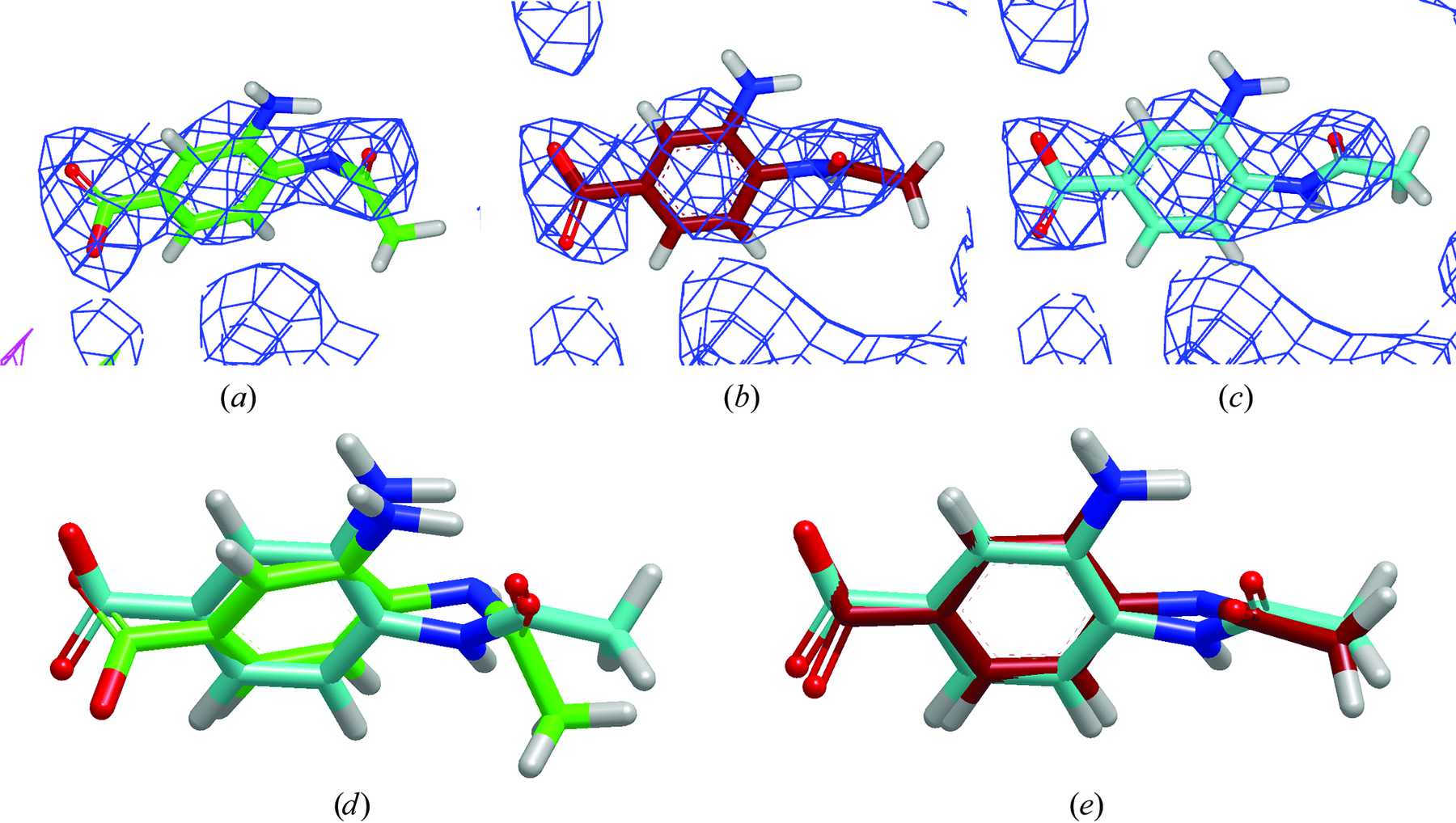 | Improved ligand geometries in crystallographic refinement using AFITT in PHENIX Acta Crystallograhpica Section D Structucal Biology (2016) 72, 1062-1072 DOI: 10.1107/S2059798316012225 PMC5013598 Modern crystal structure refinement programs rely on geometry restraints to overcome the challenge of a low data-to-parameter ratio. While the classical Engh and Huber restraints work well for standard amino-acid residues, the chemical complexity of small-molecule ligands presents a particular challenge. Most current approaches either limit ligand restraints to those that can be readily described in the Crystallographic Information File (CIF) format, thus sacrificing chemical flexibility and energetic accuracy, or they employ protocols that substantially lengthen the refinement time, potentially hindering rapid automated refinement workflows. PHENIX-AFITT refinement uses a full molecular-mechanics force field for user-selected small-molecule ligands during refinement, eliminating the potentially difficult problem of finding or generating high-quality geometry restraints. It is fully integrated with a standard refinement protocol and requires practically no additional steps from the user, making it ideal for high-throughput workflows. PHENIX-AFITT refinements also handle multiple ligands in a single model, alternate conformations and covalently bound ligands. Here, the results of combining AFITT and the PHENIX software suite on a data set of 189 protein-ligand PDB structures are presented. Refinements using PHENIX-AFITT significantly reduce ligand conformational energy and lead to improved geometries without detriment to the fit to the experimental data. For the data presented, PHENIX-AFITT refinements result in more chemically accurate models for small-molecule ligands. Read More View Full Article Download PDF |
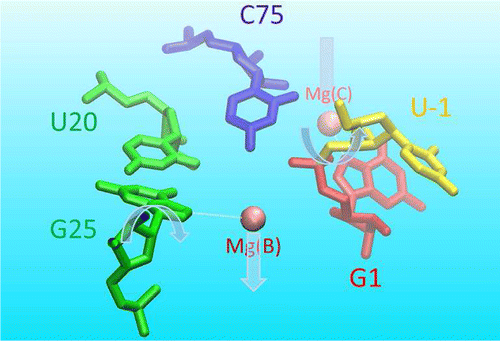 | A Two-Metal-Ion-Mediated Conformational Switching Pathway for HDV Ribozyme Activation ACS Catalysis (2016) 6, 1853-1869 DOI: 10.1021/acscatal.5b02158 PMC5072530 RNA enzymes serve as a potentially powerful platform from which to design catalysts and engineer new biotechnology. A fundamental understanding of these systems provides insight to guide design. The hepatitis delta virus ribozyme (HDVr) is a small, self-cleaving RNA motif widely distributed in nature, which has served as a paradigm for understanding the basic principles of RNA catalysis. Nevertheless, questions remain regarding the precise roles of divalent metal ions and key nucleotides in catalysis. In an effort to establish a reaction mechanism model consistent with available experimental data, we utilize molecular dynamics simulations to explore different conformations and metal ion binding modes along the HDVr reaction path. Building upon recent crystallographic data, our results provide a dynamic model of the HDVr reaction mechanism involving a conformational switch between multiple noncanonical G25:U20 base pair conformations in the active site. These local nucleobase dynamics play an important role in catalysis by modulating the metal binding environments of two Mg2+ ions that support catalysis at different steps of the reaction pathway. The first ion plays a structural role by inducing a base pair flip necessary to obtain the catalytic fold in which C75 moves towards to the scissile phosphate in the active site. Ejection of this ion then permits a second ion to bind elsewhere in the active site and facilitate nucleophile activation. The simulations collectively describe a mechanistic scenario that is consistent with currently available experimental data from crystallography, phosphorothioate substitutions, and chemical probing studies. Avenues for further experimental verification are suggested. Read More View Full Article Download PDF |
 | Creation of Academic Social Networks (ASNs) for Effective Online eLearning Communities ACS Symposium Series (2016) Chapter 9, 109-126 DOI: 10.1021/bk-2016-1217.ch009 ISBN: 9780841231252College courses with a history of large enrollment sizes, such as General Chemistry, often rely on online homework systems to provide students with practice in applying new concepts to solve problems. Online homework systems offer many potential advantages, including instant feedback to students, adaptive learning capability, and valuable data to instructors that help identify learning obstacles on-the-fly. However, there does not currently exist network infrastructure that allows a global community of online learners to leverage this wealth of data, which may be generated from different online systems, in order to facilitate synchronous interactions, enable higher cognitive skills to be exercised, and enhance team learning in cyberspace. We have recently developed a framework for the creation of a new networking paradigm to build effective online learning communities: Academic Social Networks (ASNs). The framework integrates several key components: problem template engines (PTEs) that generate questions or exercises that test specific learning objectives, a critical skills network (CSN) that established an underlying fingerprint for each problem that is generated, and a virtual classroom environment (VCE) that allows synchronous interactions to take place in order to enable problem solving and team learning in cyberspace. These components act together to create an environment where students can work problems in order to assess mastery of specific learning objectives. Mastery is tracked at various levels of difficulty that are determined by the set of required critical skills needed to solve each problem. In this way, the CSN provides the foundation for which problems can be connected to one another, mastery of learning objectives can be tracked, and specific learning pathways can be analyzed. A student struggling with a problem that is testing a specific learning objective can reach out to the ASN to connect with other students that have demonstrated mastery of that learning objective at the same difficulty level or higher, and that have a track record at effective peer-mentoring, in order to get help. Ultimately, this framework allows for the development of a tool that leverages the power of large enrollments to facilitate on-demand peer mentoring and delivery of custom instruction at scale. This work represents a significant advance in the development of novel online instructional technology that has promise to create new types of effective online learning communities that improve the quality of education. This may have a profound impact on how we connect with students enrolled in the growing massive open online courses (MOOCs) or those enrolled in large gateway courses at a university. Read More View Full Article Download PDF |
 | The importance of protonation and tautomerization in relative binding affinity prediction: a comparison of AMBER TI and Schrödinger FEP Journal of Computer-Aided Molecular Design (2016) 30, 533-539 DOI: 10.1007/s10822-016-9920-5 PMC6360336 In drug discovery, protonation states and tautomerization are easily overlooked. Through a Merck–Rutgers collaboration, this paper re-examined the initial settings and preparations for the Thermodynamic Integration (TI) calculation in AMBER Free-Energy Workflows, demonstrating the value of careful consideration of ligand protonation and tautomer state. Finally, promising results comparing AMBER TI and Schrödinger FEP+ are shown that should encourage others to explore the value of TI in routine Structure-based Drug Design. Read More View Full Article Download PDF |
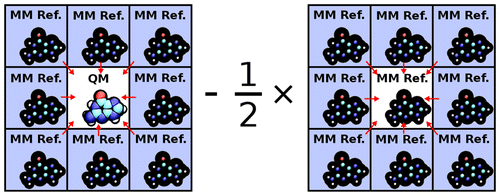 | Ambient-Potential Composite Ewald Method for ab Initio Quantum Mechanical/Molecular Mechanical Molecular Dynamics Simulation Journal of Chemical Theory and Computation (2016) 12, 2611-2632 DOI: 10.1021/acs.jctc.6b00198 PMC4907809A new approach for performing Particle Mesh Ewald in ab initio quantum mechanical/molecular mechanical (QM/MM) simulations with extended atomic orbital basis sets is presented. The new approach, the Ambient-Potential Composite Ewald (CEw) method, does not perform the QM/MM interaction with Mulliken charges nor electrostatically fit charges. Instead the nuclei and electron density interact directly with the MM environment, but in a manner that avoids the use of dense Fourier transform grids. By performing the electrostatics with the underlying QM density, the CEw method avoids self-consistent field instabilities that have been encountered with simple charge mapping procedures. Potential of mean force (PMF) profiles of the p-nitrophenyl phosphate dissociation reaction in explicit solvent are computed from PBE0/6-31G* QM/MM molecular dynamics simulations with various electrostatic protocols. The CEw profiles are shown to be stable with respect to real-space Ewald cutoff, whereas the PMFs computed from truncated and switched electrostatics produce artifacts. PBE0/6-311G**, AM1/d-PhoT, and DFTB2 QM/MM simulations are performed to generate two-dimensional PMF profiles of the phosphoryl transesterification reactions with ethoxide and phenoxide leaving groups. The semiempirical models incorrectly produce a concerted ethoxide mechanism, whereas PBE0 correctly produces a stepwise mechanism. The ab initio reaction barriers agree more closely to experiment than the semiempirical models. The failure of Mulliken-charge QM/MM-Ewald is analyzed. Read More View Full Article Download PDF |
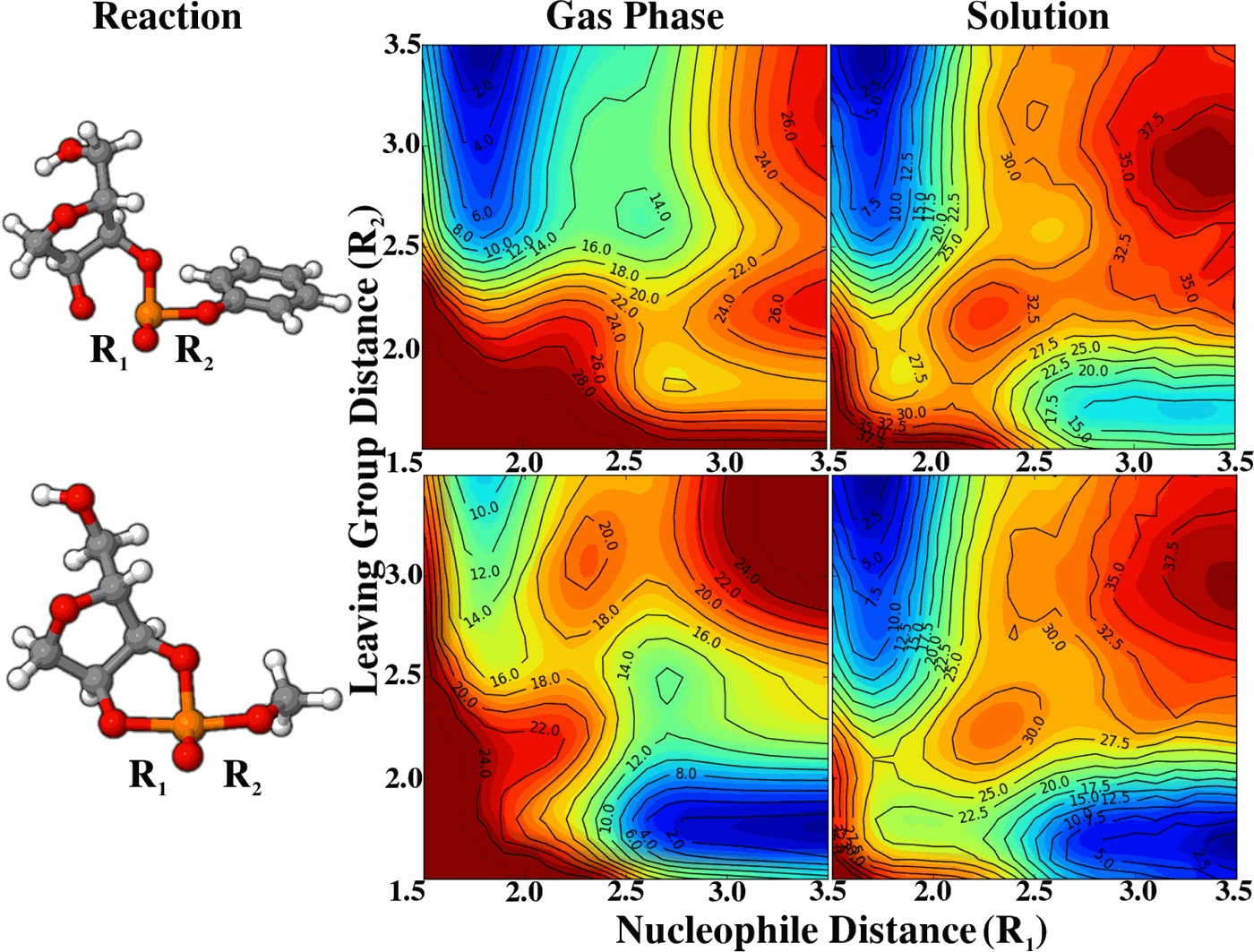 | VR-SCOSMO: A smooth conductor-like screening model with charge-dependent radii for modeling chemical reactions The Journal of Chemical Physics (2016) 144, 164115 DOI: 10.1063/1.4946779 PMC4851621To better represent the solvation effects observed along reaction pathways, and of ionic species in general, a charge-dependent variable-radii smooth conductor-like screening model (VR-SCOSMO) is developed. This model is implemented and parameterized with a third order density-functional tight binding quantum model, DFTB3/3OB-OPhyd, a quantum method which was developed for organic and biological compounds, utilizing a specific parameterization for phosphate hydrolysis reactions. Unlike most other applications with the DFTB3/3OB model, an auxiliary set of atomic multipoles is constructed from the underlying DFTB3 density matrix which is used to interact the solute with the solvent response surface. The resulting method is variational, produces smooth energies, and has analytic gradients. As a baseline, a conventional SCOSMO model with fixed radii is also parameterized. The SCOSMO and VR-SCOSMO models shown have comparable accuracy in reproducing neutral-molecule absolute solvation free energies; however, the VR-SCOSMO model is shown to reduce the mean unsigned errors (MUEs) of ionic compounds by half (about 2-3 kcal/mol). The VR-SCOSMO model presents similar accuracy as a charge-dependent Poisson-Boltzmann model introduced by Hou et al. [J. Chem. Theory Comput. 6, 2303 (2010)]. VR-SCOSMO is then used to examine the hydrolysis of trimethylphosphate and seven other phosphoryl transesterification reactions with different leaving groups. Two-dimensional energy landscapes are constructed for these reactions and calculated barriers are compared to those obtained from ab initio polarizable continuum calculations and experiment. Results of the VR-SCOSMO model are in good agreement in both cases, capturing the rate-limiting reaction barrier and the nature of the transition state. Read More View Full Article Download PDF |
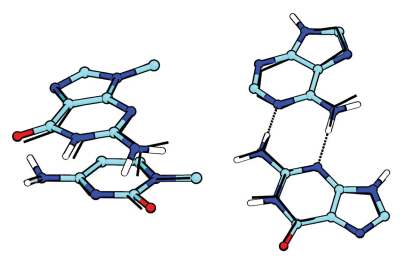 | A Modified Divide-and-Conquer Linear-Scaling Quantum Force Field with Multipolar Charge Densities Many-Body Effects and Electrostatics in Biomolecules, Pan Stanford Publishing, Singapore (2016) Chapter 2, 1-32 DOI: 10.4032/9789814613934 ISBN: 9789814613927 Recent advances in biomolecular modeling have emphasized the importance of inclusion of explicit electronic polarizabilty, and a description of electrostatic interactions that includes atomic multipoles; however, these additional levels of treatment necessarily increase a model’s computational cost. Ultimately, the decision as to whether inclusion of these more rigorous levels are justified rests on the degree to which they impact the specific application areas of interest, balanced with the overhead of their computational cost. The purpose of this book is to stimulate the exchange of effective Many-Body Effects and Electrostatics in Biomolecules strategies used to describe many-body effects and electrostatics across the quantum, classical, and coarse-grained modeling regimes Read More View Full Article Download PDF |
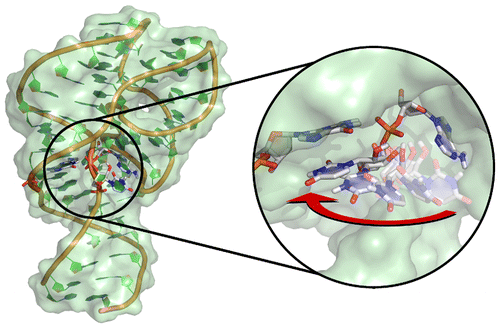 | Ribozyme catalysis with a twist: the active state of the twister ribozyme in solution predicted from molecular simulation Journal of the American Chemical Society (2016) 138, 3058-3065 DOI: 10.1021/jacs.5b12061 PMC4904722 We present results from molecular dynamics simulations and free energy calculations of the twister ribozyme at different stages along the reaction path in order to gain insight into its mechanism. Results, together with recent biochemical experiments, provide support for a mechanism involving general acid catalysis by a conserved adenine residue in the active site. Although adenine has been previously implicated as a general acid acting through the N1 position in other ribozymes such as the hairpin and VS ribozymes, in the twister ribozyme there may be - a twist. Biochemical experiments suggest that general acid catalysis may occur through the N3 position, which has never before been implicated in this role; however, there currently lacks a detailed structural model for the active state of the twister ribozyme in solution that is consistent with these and other experiments. Simulations in a crystalline environment reported here are consistent with X-ray crystallographic data, and suggest that crystal packing contacts trap the RNA in an inactive conformation with U-1 in an extruded state that is incompatible with an in-line attack to the scissile phosphate. Simulations in solution, on the other hand, reveal this region to be dynamic and able to adopt a conformation where U-1 is stacked with G33. In this state, the nucleophile is in line with the scissile phosphate, and the N1 position of G33 and N3 position of A1 are poised to act as general base and acid, respectively, as supported by mutational experiments. Free energy calculations further predict the electrostatic environment causes a shift of the microscopic pKa at the N3 position of A1 toward neutrality by approximately 5 pKa units. These results offer a unified interpretation of a broad range of currently available experimental data that points to a novel mode of general acid catalysis through the N3 position of an adenine nucleobase, thus expanding the repertoire of known mechanistic strategies employed by small nucleolytic ribozymes. Read More View Full Article Download PDF |
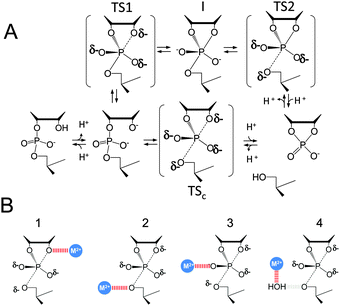 | Isotope effect analyses provide evidence for an altered transition state for RNA 2'-O-transphosphorylation catalyzed by Zn2+ Chemical Communications (2016) 52, 4462-4465 DOI: 10.1039/C5CC10212J PMC4916458 Solvent D2O and 18O kinetic isotope effects on RNA 2'-O-transphosphorylation catalyzed by Zn2+ demonstrate an altered transition state relative to specific base catalysis. A recent model from DFT calculations involving inner sphere coordination to the non-bridging and leaving group oxygens is consistent with the data. Read More View Full Article Download PDF |
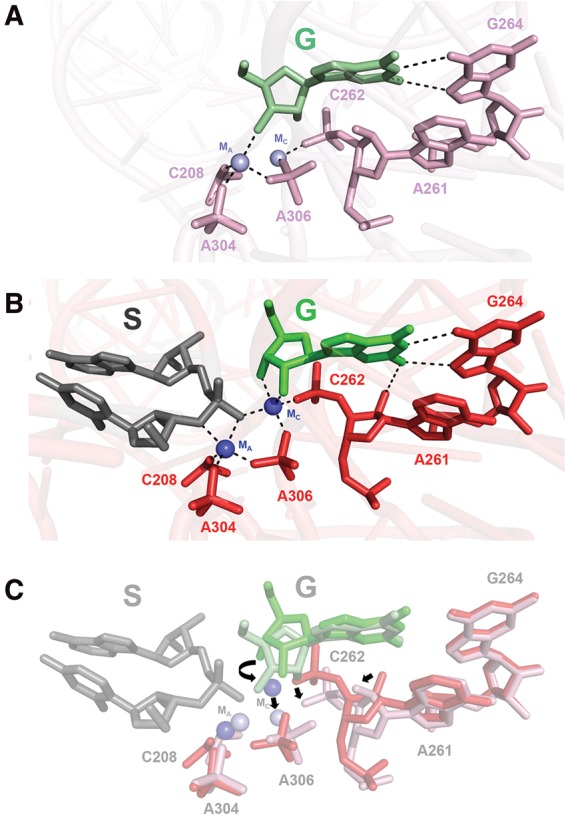 | An active site rearrangement within the Tetrahymena group I ribozyme releases nonproductive interactions and allows formation of catalytic interactions. RNA (2016) 22, 32-48 DOI: 10.1261/rna.053710.115 PMC4691833 Biological catalysis hinges on the precise structural integrity of an active site that binds and transforms its substrates and meeting this requirement presents a unique challenge for RNA enzymes. Functional RNAs, including ribozymes, fold into their active conformations within rugged energy landscapes that often contain misfolded conformers. Here we uncover and characterize one such “off-pathway” species within an active site after overall folding of the ribozyme is complete. The Tetrahymena group I ribozyme (E) catalyzes cleavage of an oligonucleotide substrate (S) by an exogenous guanosine (G) cofactor. We tested whether specific catalytic interactions with G are present in the preceding E•S•G and E•G ground-state complexes. We monitored interactions with G via the effects of 2'- and 3'-deoxy (–H) and -amino (–NH2) substitutions on G binding. These and prior results reveal that G is bound in an inactive configuration within E•G, with the nucleophilic 3'-OH making a nonproductive interaction with an active site metal ion termed MA and with the adjacent 2'-OH making no interaction. Upon S binding, a rearrangement occurs that allows both –OH groups to contact a different active site metal ion, termed MC, to make what are likely to be their catalytic interactions. The reactive phosphoryl group on S promotes this change, presumably by repositioning the metal ions with respect to G. This conformational transition demonstrates local rearrangements within an otherwise folded RNA, underscoring RNA's difficulty in specifying a unique conformation and highlighting Nature's potential to use local transitions of RNA in complex function Read More View Full Article Download PDF |
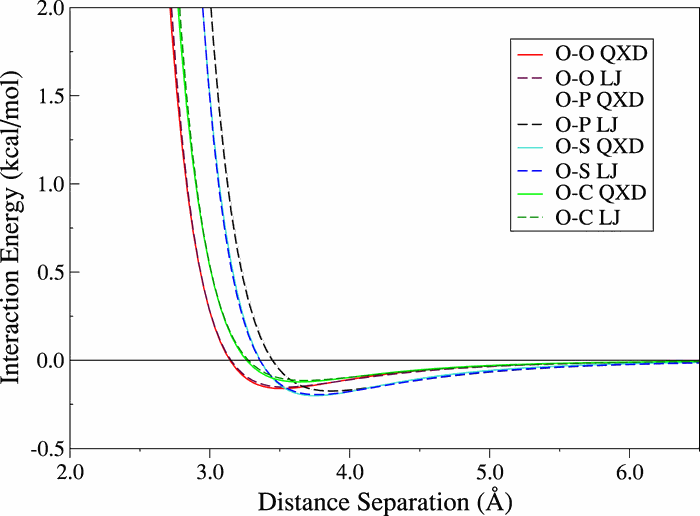 | Charge-dependent many-body exchange and dispersion interactions in combined QM/MM simulations The Journal of Chemical Physics (2015) 143, 234111 DOI: 10.1063/1.4937166 PMC4691249Accurate modeling of the molecular environment is critical in condensed phase simulations of chemical reactions. Conventional quantum mechanical/molecular mechanical (QM/MM) simulations traditionally model non-electrostatic non-bonded interactions through an empirical Lennard-Jones (LJ) potential which, in violation of intuitive chemical principles, is bereft of any explicit coupling to an atom’s local electronic structure. This oversight results in a modelwhereby short-ranged exchange-repulsion and long-ranged dispersion interactions are invariant to changes in the local atomic charge, leading to accuracy limitations for chemical reactions where significant atomic charge transfer can occur along the reaction coordinate. The present work presents a variational, charge-dependent exchange-repulsion and dispersionmodel, referred to as the charge-dependent exchange and dispersion (QXD) model, for hybrid QM/MM simulations. Analytic expressions for the energy and gradients are provided, as well as a description of the integration of the model into existing QM/MM frameworks, allowing QXD to replace traditional LJ interactions in simulations of reactive condensed phase systems. After initial validation against QM data, the method is demonstrated by capturing the solvation free energies of a series of small, chlorine-containing compounds that have varying charge on the chlorine atom. The model is further tested on the SN2 attack of a chloride anion on methylchloride. Results suggest that the QXD model, unlike the traditional LJ model, is able to simultaneously obtain accurate solvation free energies for a range of compounds while at the same time closely reproducing the experimental reactionfree energy barrier. The QXD interaction model allows explicit coupling of atomic charge with many-body exchange and dispersion interactions that are related to atomic size and provides a more accurate and robust representation of non-electrostatic non-bonded QM/MM interactions. Read More View Full Article Download PDF |
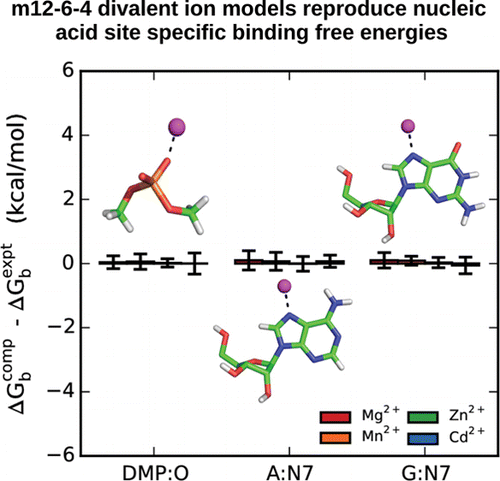 | Force Field for Mg2+, Mn2+, Zn2+, and Cd2+ Ions That Have Balanced Interactions with Nucleic Acids The Journal of Physical Chemistry B (2015) 119, 15460-15460 DOI: 10.1021/acs.jpcb.5b10423 PMC4762653 Divalent metal ions are of fundamental importance to the function and folding of nucleic acids. Divalent metal ion–nucleic acid interactions are complex in nature and include both territorial and site specific binding. Commonly employed nonbonded divalent ion models, however, are often parametrized against bulk ion properties and are subsequently utilized in biomolecular simulations without considering any data related to interactions at specific nucleic acid sites. Previously, we assessed the ability of 17 different nonbonded Mg2+ ion models to reproduce different properties of Mg2+ in aqueous solution including radial distribution functions, solvation free energies, water exchange rates, and translational diffusion coefficients. In the present work, we depart from the recently developed 12–6–4 potential models for divalent metal ions developed by Li and Merz and tune the pairwise parameters for Mg2+, Mn2+, Zn2+, and Cd2+binding dimethyl phosphate, adenosine, and guanosine in order to reproduce experimental site specific binding free energies derived from potentiometric pH titration data. We further apply these parameters to investigate a metal ion migration previously proposed to occur during the catalytic reaction of the hammerhead ribozyme. The new parameters are shown to be accurate and balanced for nucleic acid binding in comparison with available experimental data and provide an important tool for molecular dynamics and free energy simulations of nucleic acids where these ions may exhibit different binding modes. Read More View Full Article Download PDF |
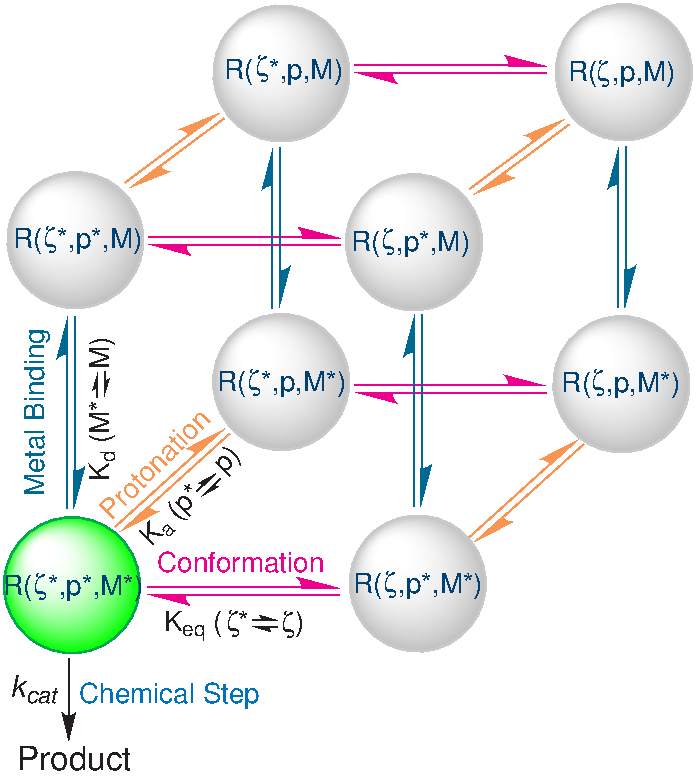 | Multiscale methods for computational RNA enzymology Methods in Enzymology (2015) 553, 335-374 DOI: 10.1016/bs.mie.2014.10.064 PMC4739856RNA catalysis is of fundamental importance to biology and yet remains ill-understood due to its complex nature. The multidimensional “problem space” of RNA catalysis includes both local and global conformational rearrangements, changes in the ion atmosphere around nucleic acids and metal ion binding, dependence on potentially correlated protonation states of key residues, and bond breaking/forming in the chemical steps of the reaction. The goal of this chapter is to summarize and apply multiscale modeling methods in an effort to target the different parts of the RNA catalysis problem space while also addressing the limitations and pitfalls of these methods. Classical molecular dynamics simulations, reference interaction site model calculations, constant pH molecular dynamics (CpHMD) simulations, Hamiltonian replica exchange molecular dynamics, and quantum mechanical/molecular mechanical simulations will be discussed in the context of the study of RNA backbone cleavage transesterification. This reaction is catalyzed by both RNA and protein enzymes, and here we examine the different mechanistic strategies taken by the hepatitis delta virus ribozyme and RNase A. Read More View Full Article Download PDF |
 | Multipolar Ewald Methods. II. Applications Using a Quantum Mechanical Force Field Journal of Chemical Theory and Computation (2015) 11, 451-461 DOI: 10.1021/ct500799g PMC4325604 A fully quantum mechanical force field (QMFF) based on a modified “divide-and-conquer” (mDC) framework is applied to a series of molecular simulation applications, using a generalized Particle Mesh Ewald method extended to multipolar charge densities. Simulation results are presented for three example applications: liquid water, p-nitrophenylphosphate reactivity in solution, and crystalline N,N-dimethylglycine. Simulations of liquid water using a parametrized mDC model are compared to TIP3P and TIP4P/Ew water models and experiment. The mDC model is shown to be superior for cluster binding energies and generally comparable for bulk properties. Examination of the dissociative pathway for dephosphorylation of p-nitrophenylphosphate shows that the mDC method evaluated with the DFTB3/3OB and DFTB3/OPhyd semiempirical models bracket the experimental barrier, whereas DFTB2 and AM1/d-PhoT QM/MM simulations exhibit deficiencies in the barriers, the latter for which is related, in part, to the anomalous underestimation of the p-nitrophenylate leaving group pKa. Simulations of crystalline N,N-dimethylglycine are performed and the overall structure and atomic fluctuations are compared with the experiment and the general AMBER force field (GAFF). The QMFF, which was not parametrized for this application, was shown to be in better agreement with crystallographic data than GAFF. Our simulations highlight some of the application areas that may benefit from using new QMFFs, and they demonstrate progress toward the development of accurate QMFFs using the recently developed mDC framework. Read More View Full Article Download PDF |
 | Multipolar Ewald Methods. I. Theory, Accuracy, and Performance Journal of Chemical Theory and Computation (2015) 11, 436-450 DOI: 10.1021/ct5007983 PMC4325605The Ewald, Particle Mesh Ewald (PME), and Fast Fourier–Poisson (FFP) methods are developed for systems composed of spherical multipole moment expansions. A unified set of equations is derived that takes advantage of a spherical tensor gradient operator formalism in both real space and reciprocal space to allow extension to arbitrary multipole order. The implementation of these methods into a novel linear-scaling modified “divide-and-conquer” (mDC) quantum mechanical force field is discussed. The evaluation times and relative force errors are compared between the three methods, as a function of multipole expansion order. Timings and errors are also compared within the context of the quantum mechanical force field, which encounters primary errors related to the quality of reproducing electrostatic forces for a given density matrix and secondary errors resulting from the propagation of the approximate electrostatics into the self-consistent field procedure, which yields a converged, variational, but nonetheless approximate density matrix. Condensed-phase simulations of an mDC water model are performed with the multipolar PME method and compared to an electrostatic cutoff method, which is shown to artificially increase the density of water and heat of vaporization relative to full electrostatic treatment. Read More View Full Article Download PDF |
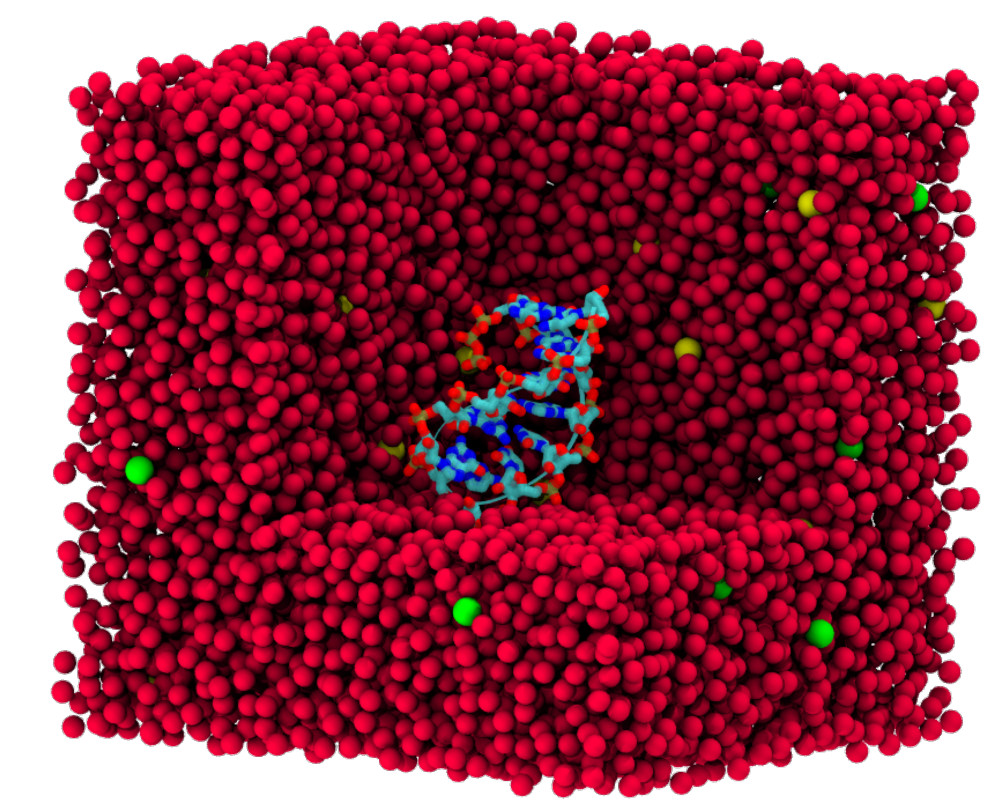 | Structural fidelity and NMR relaxation analysis in a prototype RNA hairpin RNA (2015) 21, 963-974 DOI: 10.1261/rna.047357.114 PMC4408802RNA hairpins are widespread and very stable motifs that contribute decisively to RNA folding and biological function. The GTP1G2C3A4C5U6U7C8G9G10U11G12C13C14 construct (with a central UUCG tetraloop) has been extensively studied by solution NMR, and offers and excellent opportunity to evaluate the structure and dynamical description afforded by molecular dynamics (MD) simulations. Here, we compare average structural parameters and NMR relaxation rates estimated from a series of multiple independent explicit solvent MD simulations using the two most recent RNA AMBER force fields (ff99 and ff10). Predicted overall tumbling times are ~20% faster than those inferred from analysis of NMR data and follow the same trend when temperature and ionic strength is varied. The Watson–Crick stem and the “canonical” UUCG loop structure is maintained in most simulations including the characteristic syn conformation along the glycosidic bond of G9, although some key hydrogen bonds in the loop are partially disrupted. Our analysis pinpoints G9–G10 backbone conformations as a locus of discrepancies between experiment and simulation. In general the results for the more recent force-field parameters (ff10) are closer to experiment than those for the older ones (ff99). This work provides a comprehensive and detailed comparison of state of the art MD simulations against a wide variety of solution NMR measurements. Read More View Full Article Download PDF |
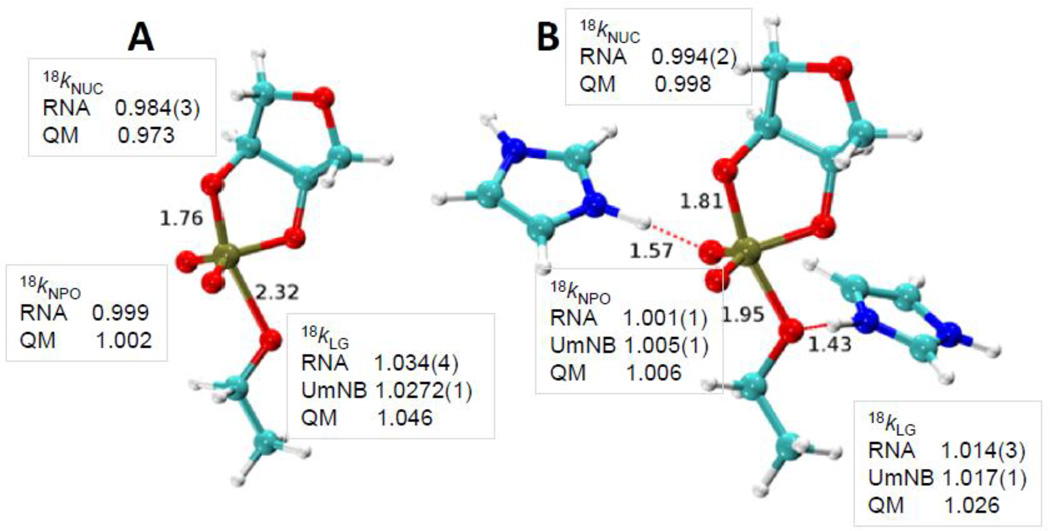 | Integration of kinetic isotope effect analyses to elucidate ribonuclease mechanism Biochimica et Biophysica Acta (2015) 1854, 1801-1808 DOI: 10.1016/j.bbapap.2015.04.022 PMC4604014 The well-studied mechanism of ribonuclease A is believed to involve concerted general acid-base catalysis by two histidine residues, His12 and His119. The basic features of this mechanism are often cited to explain rate enhancement by both protein and RNA enzymes that catalyze RNA 2'-O-transphosphorylation. Recent kinetic isotope effect analyses and computational studies are providing a more chemically detailed description of the mechanism of RNase A and the rate limiting transition state. Overall, the results support an asynchronous mechanism for both solution and ribonuclease catalyzed reactions in which breakdown of a transient dianoinic phosphorane intermediate by 5'OP bond cleavage is rate limiting. Relative to non-enzymatic reactions catalyzed by specific base, a smaller KIE on the 5'O leaving group and a less negative ßLG are observed for RNase A catalysis. Quantum mechanical calculations consistent with these data support a model in which electrostatic and H-bonding interactions with the non-bridging oxygens and proton transfer from His119 render departure of the 5'O less advanced and stabilize charge buildup in the transition state. Both experiment and computation indicate advanced 2'OP bond formation in the rate limiting transition state. However, this feature makes it difficult to resolve the chemical steps involved in 2'O activation. Thus, modeling the transition state for RNase A catalysis underscores those elements of its chemical mechanism that are well resolved, as well as highlighting those where ambiguity remains. This article is part of a Special Issue entitled: Enzyme Transition States from Theory and Experiment. Read More View Full Article Download PDF |
 | Enzyme transition states from theory and experiment Biochimica et Biophysica Acta (2015) 1854, 1727-1728 DOI: 10.1016/j.bbapap.2015.08.006 26302659 Understanding how enzymes catalyze specific reactions has yielded important insights into biology, provided the necessary context for the design of novel catalysts, and has facilitated the development of enzyme inhibitors for therapeutic purposes. The contributions of Professor W. Wallace Cleland helped to forge the modern framework for steady state enzyme kinetics, and together with his colleagues he made pioneering contributions to establishing heavy atom isotope effect analyses as a powerful approach for investigating the chemical mechanisms of enzyme-catalyzed reactions. Professor Cleland collaborated with and trained numerous talented enzymologists and generations of scientists. Several facets of W.W. Cleland's contributions to science are illustrated and reflected in this special issue that focuses on using kinetic isotope effects to understand chemical and enzymatic reaction mechanisms. Read More View Full Article Download PDF |
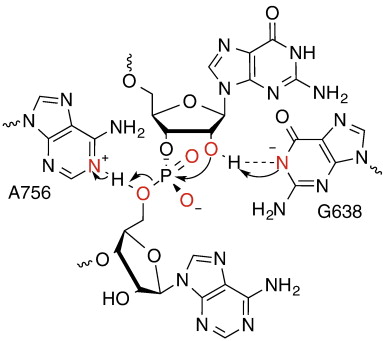 | Heavy Atom Labeled Nucleotides for Measurement of Kinetic Isotope Effects Biochimica et Biophysica Acta (2015) 1854, 1737-1745 DOI: 10.1016/j.bbapap.2015.03.007 PMC4584197 Experimental analysis of kinetic isotope effects represents an extremely powerful approach for gaining information about the transition state structure of complex reactions not available through other methodologies. The implementation of this approach to the study of nucleic acid chemistry requires the synthesis of nucleobases and nucleotides enriched for heavy isotopes at specific positions. In this review, we highlight current approaches to the synthesis of nucleic acids enriched site specifically for heavy oxygen and nitrogen and their application in heavy atom isotope effect studies. This article is part of a special issue titled: Enzyme Transition States from Theory and Experiment. Read More View Full Article Download PDF |
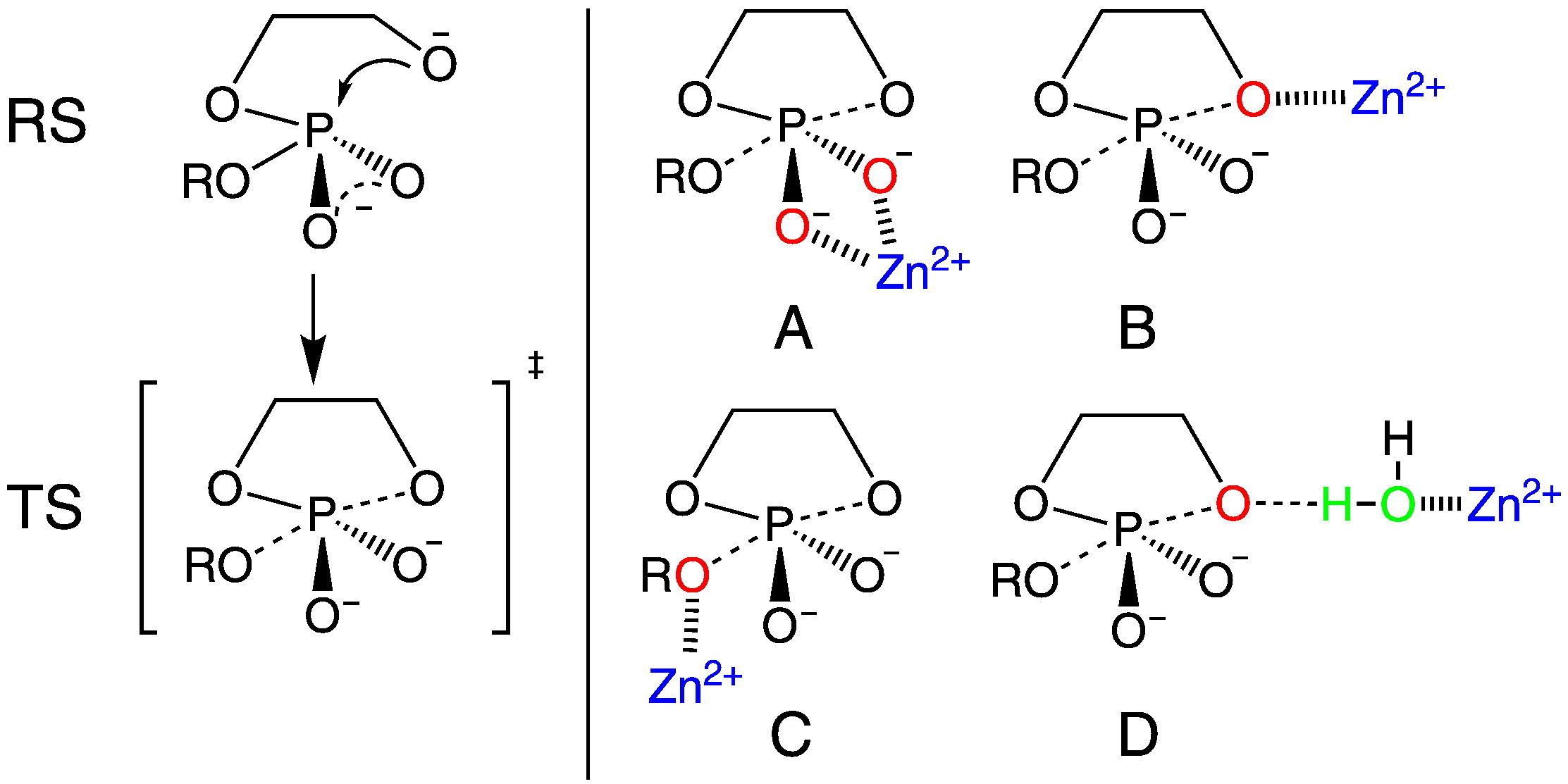 | Effect of Zn2+ binding and enzyme active site on the transition state for RNA 2'-O-transphosphorylation interpreted through kinetic isotope effects Biochimica et Biophysica Acta (2015) 1854, 1795-1800 DOI: 10.1016/j.bbapap.2015.02.022. PMC4743498 Divalent metal ions, due to their ability to stabilize high concentrations of negative charge, are important for RNA folding and catalysis. Detailed models derived from the structures and kinetics of enzymes and from computational simulations have been developed. However, in most cases the specific catalytic modes involving metal ions and their mechanistic roles and effects on transition state structures remain controversial. Valuable information about the nature of the transition state is provided by measurement of kinetic isotope effects (KIEs). However, KIEs reflect changes in all bond vibrational modes that differ between the ground state and transition state. QM calculations are therefore essential for developing structural models of the transition state and evaluating mechanistic alternatives. Herein, we present computational models for Zn2+ binding to RNA 2'O-transphosphorylation reaction models that aid in the interpretation of KIE experiments. Different Zn2+ binding modes produce distinct KIE signatures, and one binding mode involving two zinc ions is in close agreement with KIEs measured for non-enzymatic catalysis by Zn2+ aquo ions alone. Interestingly, the KIE signatures in this specific model are also very close to those in RNase A catalysis. These results allow a quantitative connection to be made between experimental KIE measurements and transition state structure and bonding, and provide insight into RNA 2'O-ransphosphorylation reactions catalyzed by metal ions and enzymes. This article is part of a Special Issue entitled: Enzyme Transition States from Theory and Experiment. Read More View Full Article Download PDF |
 | Cation–Anion Interactions within the Nucleic Acid Ion Atmosphere Revealed by Ion Counting Journal of the American Chemical Society (2015) 137, 14705-14715 DOI: 10.1021/jacs.5b08395 PMC4739826The ion atmosphere is a critical structural, dynamic, and energetic component of nucleic acids that profoundly affects their interactions with proteins and ligands. Experimental methods that "count" the number of ions thermodynamically associated with the ion atmosphere allow dissection of energetic properties of the ion atmosphere, and thus provide direct comparison to theoretical results. Previous experiments have focused primarily on the cations that are attracted to nucleic acid polyanions, but have also showed that anions are excluded from the ion atmosphere. Herein, we have systematically explored the properties of anion exclusion, testing the zeroth-order model that anions of different identity are equally excluded due to electrostatic repulsion. Using a series of monovalent salts, we find, surprisingly, that the extent of anion exclusion and cation inclusion significantly depends on salt identity. The differences are prominent at higher concentrations and mirror trends in mean activity coefficients of the electrolyte solutions. Salts with lower activity coefficients exhibit greater accumulation of both cations and anions within the ion atmosphere, strongly suggesting that cation–anion correlation effects are present in the ion atmosphere and need to be accounted for to understand electrostatic interactions of nucleic acids. To test whether the effects of cation–anion correlations extend to nucleic acid kinetics and thermodynamics, we followed the folding of P4–P6, a domain of the Tetrahymena group I ribozyme, via single-molecule fluorescence resonance energy transfer in solutions with different salts. Solutions of identical concentration but lower activity gave slower and less favorable folding. Our results reveal hitherto unknown properties of the ion atmosphere and suggest possible roles of oriented ion pairs or anion-bridged cations in the ion atmosphere for electrolyte solutions of salts with reduced activity. Consideration of these new results leads to a reevaluation of the strengths and limitations of Poisson–Boltzmann theory and highlights the need for next-generation atomic-level models of the ion atmosphere. Read More View Full Article Download PDF |
 | Competitive interaction of monovalent cations with DNA from 3D-RISM Nucleic Acids Research (2015) 43, 8405-8415 DOI: 10.1093/nar/gkv830 PMC4787805 The composition of the ion atmosphere surrounding nucleic acids affects their folding, condensation and binding to other molecules. It is thus of fundamental importance to gain predictive insight into the formation of the ion atmosphere and thermodynamic consequences when varying ionic conditions. An early step toward this goal is to benchmark computational models against quantitative experimental measurements. Herein, we test the ability of the three dimensional reference interaction site model (3D-RISM) to reproduce preferential interaction parameters determined from ion counting (IC) experiments for mixed alkali chlorides and dsDNA. Calculations agree well with experiment with slight deviations for salt concentrations >200 mM and capture the observed trend where the extent of cation accumulation around the DNA varies inversely with its ionic size. Ion distributions indicate that the smaller, more competitive cations accumulate to a greater extent near the phosphoryl groups, penetrating deeper into the grooves. In accord with experiment, calculated IC profiles do not vary with sequence, although the predicted ion distributions in the grooves are sequence and ion size dependent. Calculations on other nucleic acid conformations predict that the variation in linear charge density has a minor effect on the extent of cation competition. Read More View Full Article Download PDF |
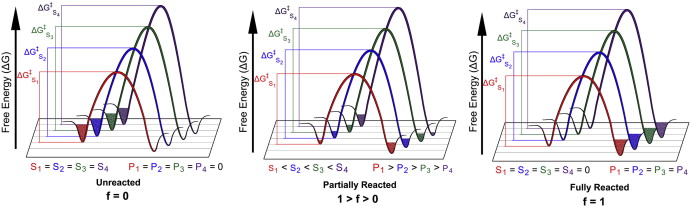 | Determination of hepatitis delta virus ribozyme N(–1) nucleobase and functional group specificity using internal competition kinetics Analytical Biochemistry (2015) 483, 12-20 DOI: 10.1016/j.ab.2015.04.024 PMC4461535Biological catalysis involves interactions distant from the site of chemistry that can position the substrate for reaction. Catalysis of RNA 2'-O-transphosphorylation by the hepatitis delta virus (HDV) ribozyme is sensitive to the identity of the N(–1) nucleotide flanking the reactive phosphoryl group. However, the interactions that affect the conformation of this position, and in turn the 2'O nucleophile, are unclear. Here, we describe the application of multiple substrate internal competition kinetic analyses to understand how the N(–1) nucleobase contributes to HDV catalysis and test the utility of this approach for RNA structure–function studies. Internal competition reactions containing all four substrate sequence variants at the N(–1) position in reactions using ribozyme active site mutations at A77 and A78 were used to test a proposed base-pairing interaction. Mutants A78U, A78G, and A79G retain significant catalytic activity but do not alter the specificity for the N(–1) nucleobase. Effects of nucleobase analog substitutions at N(–1) indicate that U is preferred due to the ability to donate an H-bond in the Watson–Crick face and avoid minor groove steric clash. The results provide information essential for evaluating models of the HDV active site and illustrate multiple substrate kinetic analyses as a practical approach for characterizing structure–function relationships in RNA reactions. Read More View Full Article Download PDF |
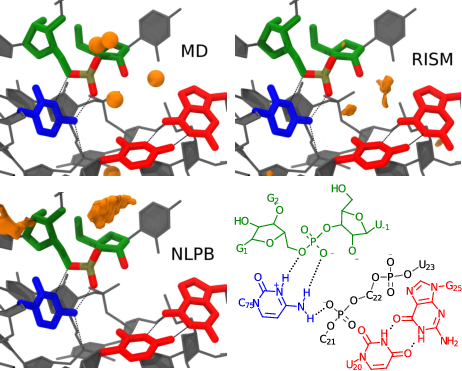 | Assessment of Metal-Assisted Nucleophile Activation in the Hepatitis Delta Virus Ribozyme from Molecular Simulation and 3D-RISM RNA (2015) 21, 1-12 DOI: 10.1261/rna.051466.115 PMC4536318 The hepatitis delta virus ribozyme is an efficient catalyst of RNA 2'-O-transphosphorylation and has emerged as a key experimental system for identifying and characterizing fundamental features of RNA catalysis. Recent structural and biochemical data have led to a proposed mechanistic model whereby an active site Mg2+ ion facilitates deprotonation of the O2' nucleophile, and a protonated cytosine residue (C75) acts as an acid to donate a proton to the O5' leaving group as noted in a previous study. This model assumes that the active site Mg2+ ion forms an inner-sphere coordination with the O2' nucleophile and a nonbridging oxygen of the scissile phosphate. These contacts, however, are not fully resolved in the crystal structure, and biochemical data are not able to unambiguously exclude other mechanistic models. In order to explore the feasibility of this model, we exhaustively mapped the free energy surfaces with different active site ion occupancies via quantum mechanical/molecular mechanical (QM/MM) simulations. We further incorporate a three-dimensional reference interaction site model for the solvated ion atmosphere that allows these calculations to consider not only the rate associated with the chemical steps, but also the probability of observing the system in the presumed active state with the Mg2+ ion bound. The QM/MM results predict that a pathway involving metal-assisted nucleophile activation is feasible based on the rate-controlling transition state barrier departing from the presumed metal-bound active state. However, QM/MM results for a similar pathway in the absence of Mg2+ are not consistent with experimental data, suggesting that a structural model in which the crystallographically determined Mg2+ is simply replaced with Na+ is likely incorrect. It should be emphasized, however, that these results hinge upon the assumption of the validity of the presumed Mg2+-bound starting state, which has not yet been definitively verified experimentally, nor explored in depth computationally. Thus, further experimental and theoretical study is needed such that a consensus view of the catalytic mechanism emerges. Read More View Full Article Download PDF |
 | Nucleic acid reactivity: challenges for next-generation semiempirical quantum models Journal of Computational Chemistry (2015) 36, 1370-1389 DOI: 10.1002/jcc.23933 PMC4760688Semiempirical quantum models are routinely used to study mechanisms of RNA catalysis and phosphoryl transfer reactions using combined quantum mechanical (QM)/molecular mechanical methods. Herein, we provide a broad assessment of the performance of existing semiempirical quantum models to describe nucleic acid structure and reactivity to quantify their limitations and guide the development of next-generation quantum models with improved accuracy. Neglect of diatomic differential overlap and self-consistent density-functional tight-binding semiempirical models are evaluated against high-level QM benchmark calculations for seven biologically important datasets. The datasets include: proton affinities, polarizabilities, nucleobase dimer interactions, dimethyl phosphate anion, nucleoside sugar and glycosidic torsion conformations, and RNA phosphoryl transfer model reactions. As an additional baseline, comparisons are made with several commonly used density-functional models, including M062X and B3LYP (in some cases with dispersion corrections). The results show that, among the semiempirical models examined, the AM1/d-PhoT model is the most robust at predicting proton affinities. AM1/d-PhoT and DFTB3-3ob/OPhyd reproduce the MP2 potential energy surfaces of 6 associative RNA phosphoryl transfer model reactions reasonably well. Further, a recently developed linear-scaling “modified divide-and-conquer” model exhibits the most accurate results for binding energies of both hydrogen bonded and stacked nucleobase dimers. The semiempirical models considered here are shown to underestimate the isotropic polarizabilities of neutral molecules by approximately 30%. The semiempirical models also fail to adequately describe torsion profiles for the dimethyl phosphate anion, the nucleoside sugar ring puckers, and the rotations about the nucleoside glycosidic bond. The modeling of pentavalent phosphorus, particularly with thio substitutions often used experimentally as mechanistic probes, was problematic for all of the models considered. Analysis of the strengths and weakness of the models suggests that the creation of robust next-generation models should emphasize the improvement of relative conformational energies and barriers, and nonbonded interactions. Read More View Full Article Download PDF |
 | Comparison of Structural, Thermodynamic, Kinetic and Mass Transport Properties of Mg2+ Models Commonly Used in Biomolecular Simulations Journal of Computational Chemistry (2015) 36, 970-982 DOI: 10.1002/jcc.23881 PMC4409555The prevalence of Mg2+ ions in biology and their essential role in nucleic acid structure and function has motivated the development of various Mg2+ ion models for use in molecular simulations. Currently, the most widely used models in biomolecular simulations represent a nonbonded metal ion as an ion-centered point charge surrounded by a nonelectrostatic pairwise potential that takes into account dispersion interactions and exchange effects that give rise to the ion's excluded volume. One strategy toward developing improved models for biomolecular simulations is to first identify a Mg2+ model that is consistent with the simulation force fields that closely reproduces a range of properties in aqueous solution, and then, in a second step, balance the ion–water and ion–solute interactions by tuning parameters in a pairwise fashion where necessary. The present work addresses the first step in which we compare 17 different nonbonded single-site Mg2+ ion models with respect to their ability to simultaneously reproduce structural, thermodynamic, kinetic and mass transport properties in aqueous solution. None of the models based on a 12-6 nonelectrostatic nonbonded potential was able to reproduce the experimental radial distribution function, solvation free energy, exchange barrier and diffusion constant. The models based on a 12-6-4 potential offered improvement, and one model in particular, in conjunction with the SPC/E water model, performed exceptionally well for all properties. The results reported here establish useful benchmark calculations for Mg2+ ion models that provide insight into the origin of the behavior in aqueous solution, and may aid in the development of next-generation models that target specific binding sites in biomolecules. Read More View Full Article Download PDF |
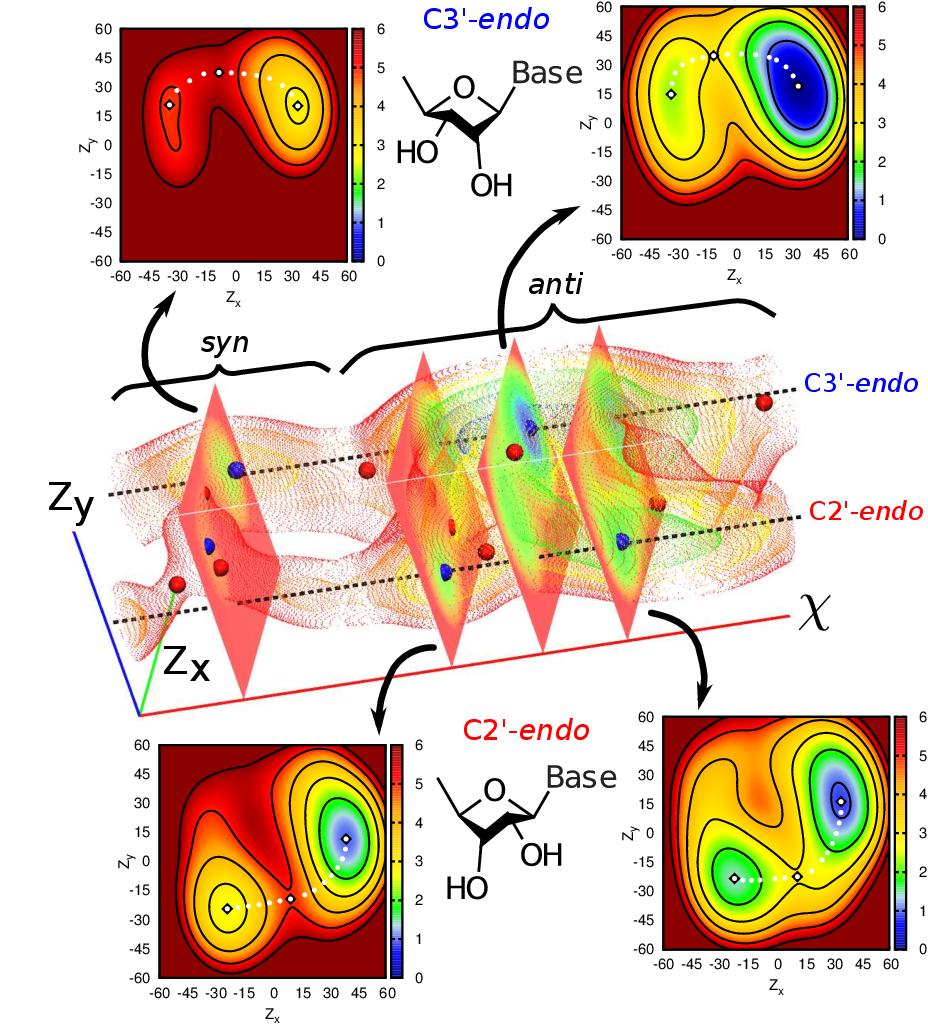 | Characterization of the three-dimensional free energy manifold for the uracil ribonucleoside from asynchronous replica exchange simulations Journal of Chemical Theory and Computation (2015) 11, 373-377 DOI: 10.1021/ct500776j PMC4745604Replica exchange molecular dynamics has emerged as a powerful tool for efficiently sampling free energy landscapes for conformational and chemical transitions. However, daunting challenges remain in efficiently getting such simulations to scale to the very large number of replicas required to address problems in state spaces beyond two dimensions. The development of enabling technology to carry out such simulations is in its infancy, and thus it remains an open question as to which applications demand extension into higher dimensions. In the present work, we explore this problem space by applying asynchronous Hamiltonian replica exchange molecular dynamics with a combined quantum mechanical/molecular mechanical potential to explore the conformational space for a simple ribonucleoside. This is done using a newly developed software framework capable of executing >3,000 replicas with only enough resources to run 2,000 simultaneously. This may not be possible with traditional synchronous replica exchange approaches. Our results demonstrate 1.) the necessity of high dimensional sampling simulations for biological systems, even as simple as a single ribonucleoside, and 2.) the utility of asynchronous exchange protocols in managing simultaneous resource requirements expected in high dimensional sampling simulations. It is expected that more complicated systems will only increase in computational demand and complexity, and thus the reported asynchronous approach may be increasingly beneficial in order to make such applications available to a broad range of computational scientists. Read More View Full Article Download PDF |
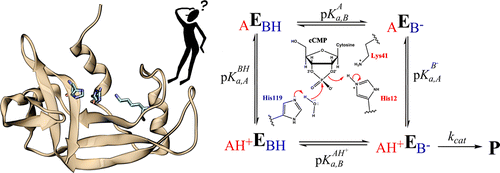 | Interpretation of pH–Activity Profiles for Acid–Base Catalysis from Molecular Simulations Biochemistry (2015) 54, 1307-1313 DOI: 10.1021/bi5012833 PMC4441796The measurement of reaction rate as a function of pH provides essential information about mechanism. These rates are sensitive to the pKa values of amino acids directly involved in catalysis that are often shifted by the enzyme active site environment. Experimentally observed pH-rate profilesare usually interpreted using simple kinetic models that allow estimation of "apparent pKa" values of presumed general acid and base catalysts. One of the underlying assumptions in these models is that the protonation states are uncorrelated. In this work, we introduce the use of constant pHmolecular dynamics simulations in explicit solvent (CpHMD) with replica exchange in the pH-dimension (pH-REMD) as a tool to aid in theinterpretation of pH-activity data of enzymes and to test the validity of different kinetic models. We apply the methods to RNase A, a prototype acid-base catalyst, to predict the macroscopic and microscopic pKa values, as well as the shape of the pH-rate profile. Results for apo and cCMP-bound RNase A agree well with available experimental data and suggest that deprotonation of the general acid and protonation of the general base are not strongly coupled in transphosphorylation and hydrolysis steps. Stronger coupling, however, is predicted for the Lys41 and His119 protonation states in apo RNase A, leading to the requirement for a microscopic kinetic model. This type of analysis may be important for other catalytic systems where the active forms of the implicated general acid and base are oppositely charged and more highly correlated. These results suggest a new way for CpHMD/pH-REMD simulations to bridge the gap with experiments to provide a molecular-level interpretation of pH-activity data in studies of enzyme mechanisms. Read More View Full Article Download PDF |
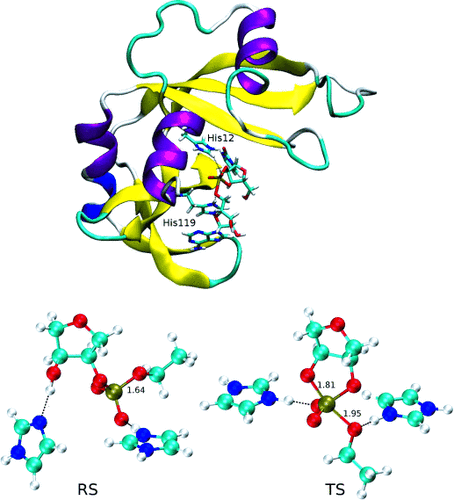 | Mechanistic insights into RNA transphosphorylation from kinetic isotope effects and linear free energy relationships of model reactions Chemistry-A European Journal (2014) 20, 1-9 DOI: 10.1002/chem.201403862 PMC4432475Phosphoryl transfer reactions are ubiquitous in biology and the understanding of the mechanisms whereby these reactions are catalyzed by protein and RNA enzymes is central to reveal design principles for new therapeutics. Two of the most powerful experimental probes of chemical mechanism involve the analysis of linear free energy relations (LFERs) and the measurement of kinetic isotope effects (KIEs). These experimental data report directly on differences in bonding between the ground state and the rate-controlling transition state, which is the most critical point along the reaction free energy pathway. However, interpretation of LFER and KIE data in terms of transition-state structure and bonding optimally requires the use of theoretical models. In this work, we apply density-functional calculations to determine KIEs for a series of phosphoryl transfer reactions of direct relevance to the 2'-O-transphosphorylation that leads to cleavage of the phosphodiester backbone of RNA. We first examine a well-studied series of phosphate and phosphorothioate mono-, di- and triesters that are useful as mechanistic probes and for which KIEs have been measured. Close agreement is demonstrated between the calculated and measured KIEs, establishing the reliability of our quantum model calculations. Next, we examine a series of RNA transesterification model reactions with a wide range of leaving groups in order to provide a direct connection between observed Brønsted coefficients and KIEs with the structure and bonding in the transition state. These relations can be used for prediction or to aid in the interpretation of experimental data for similar non-enzymatic and enzymatic reactions. Finally, we apply these relations to RNA phosphoryl transfer catalyzed by ribonuclease A, and demonstrate the reaction coordinate–KIE correlation is reasonably preserved. A prediction of the secondary deuterium KIE in this reaction is also provided. These results demonstrate the utility of building up knowledge of mechanism through the systematic study of model systems to provide insight into more complex biological systems such as phosphoryl transfer enzymes and ribozymes. Read More View Full Article Download PDF |
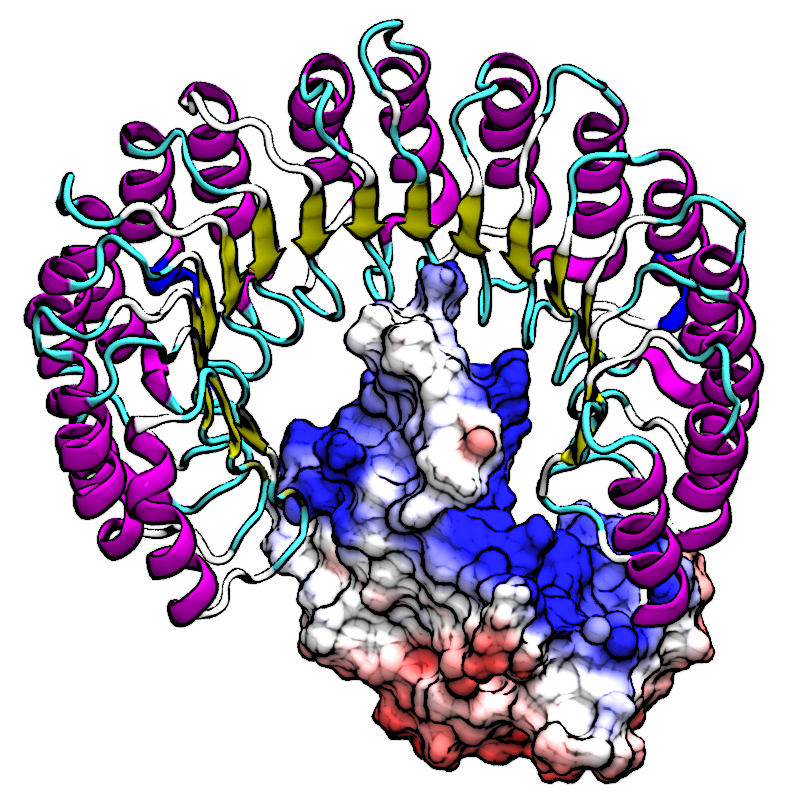 | Recent advances toward a general purpose linear-scaling quantum force field Accounts of Chemical Research (2014) 47, 2812-2820 DOI: 10.1021/ar500103g PMC4165466There is need in the molecular simulation community to develop new quantum mechanical (QM) methods that can be routinely applied to the simulation of large molecular systems in complex, heterogeneous condensed phase environments. Although conventional methods, such as the hybrid quantum mechanical/molecular mechanical (QM/MM) method, are adequate for many problems, there remain other applications that demand a fully quantum mechanical approach. QM methods are generally required in applications that involve changes in electronic structure, such as when chemical bond formation or cleavage occurs, when molecules respond to one another through polarization or charge transfer, or when matter interacts with electromagnetic fields. A full QM treatment, rather than QM/MM, is necessary when these features present themselves over a wide spatial range that, in some cases, may span the entire system. Specific examples include the study of catalytic events that involve delocalized changes in chemical bonds, charge transfer, or extensive polarization of the macromolecular environment; drug discovery applications, where the wide range of nonstandard residues and protonation states are challenging to model with purely empirical MM force fields; and the interpretation of spectroscopic observables. Unfortunately, the enormous computational cost of conventional QM methods limit their practical application to small systems. Linear-scaling electronic structure methods (LSQMs) make possible the calculation of large systems but are still too computationally intensive to be applied with the degree of configurational sampling often required to make meaningful comparison with experiment. In this work, we present advances in the development of a quantum mechanical force field (QMFF) suitable for application to biological macromolecules and condensed phase simulations. QMFFs leverage the benefits provided by the LSQM and QM/MM approaches to produce a fully QM method that is able to simultaneously achieve very high accuracy and efficiency. The efficiency of the QMFF is made possible by partitioning the system into fragments and self-consistently solving for the fragment-localized molecular orbitals in the presence of the other fragment’s electron densities. Unlike a LSQM, the QMFF introduces empirical parameters that are tuned to obtain very accurate intermolecular forces. The speed and accuracy of our QMFF is demonstrated through a series of examples ranging from small molecule clusters to condensed phase simulation, and applications to drug docking and protein–protein interactions. In these examples, comparisons are made to conventional molecular mechanical models, semiempirical methods, ab initio Hamiltonians, and a hybrid QM/MM method. The comparisons demonstrate the superior accuracy of our QMFF relative to the other models; nonetheless, we stress that the overarching role of QMFFs is not to supplant these established computational methods for problems where their use is appropriate. The role of QMFFs within the toolbox of multiscale modeling methods is to extend the range of applications to include problems that demand a fully quantum mechanical treatment of a large system with extensive configurational sampling. Read More View Full Article Download PDF |
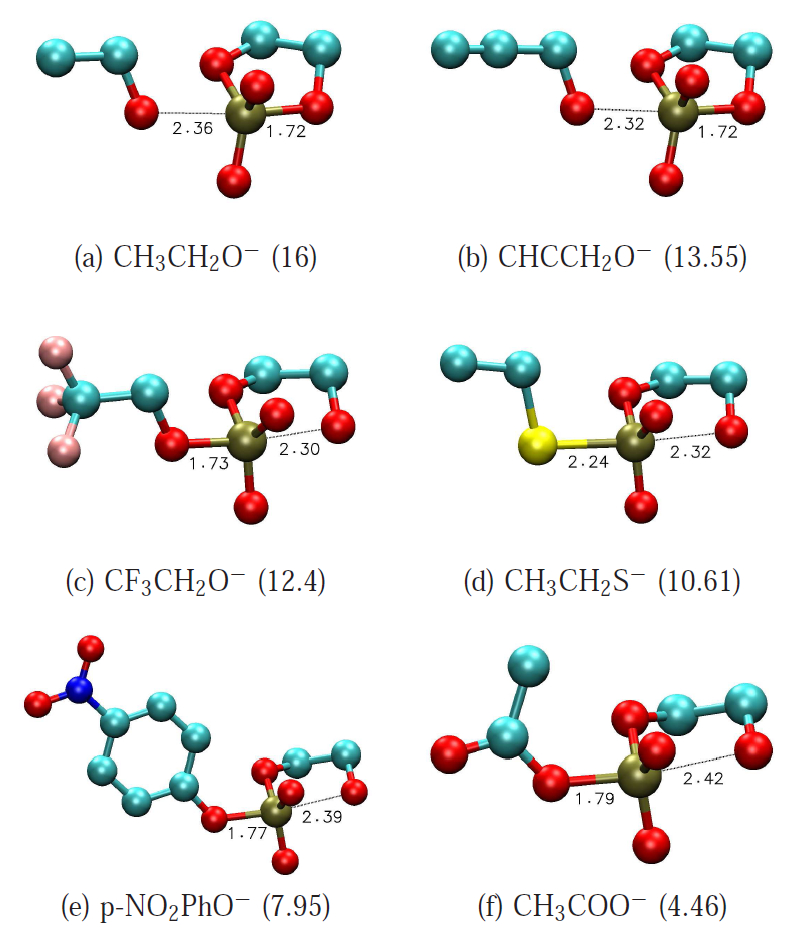 | Linear Free Energy Relationships in RNA Transesterification: Theoretical Models to Aid Experimental Interpretations Physical Chemistry Chemical Physics (2014) 16, 15846-15855 DOI: 10.1039/c4cp01050g PMC4366550RNA cleavage transesterification is of fundamental reaction in biology that is catalyzed by both protein and RNA enzymes. In this work, a series of RNA transesterification model reactions with a wide range of leaving groups are investigated with density-functional calculations in an aqueous solvation environment in order to study linear free energy relationships (LFERs) and their connection to transition state structure and bonding. Overall, results obtained from the polarizable continuum solvation model with UAKS radii produce the best linear correlations and closest overall agreement with experimental results. Reactions with a poor leaving group are predicted to proceed via a stepwise mechanism with a late transition state that is rate controlling. As leaving group becomes more acidic and labile, the barriers of both early and late transition states decrease. LFERs for each transition state are computed, with the late transition state barrier showing greater sensitivity to leaving group pKa. For sufficiently enhanced leaving groups, the reaction mechanism transits to a concerted mechanism characterized by a single early transition state. Further linear relationships were derived for bond lengths and bond orders as a function of leaving group pKa and rate constant values that can be used for prediction. This work provides important benchmark linear free energy data that allows a molecular-level characterization of the structure and bonding of the transition states for this important class of phosphoryl transfer reactions. The relations reported herein can be used to aid in the interpretation of data obtained from experimental studies of non-catalytic and catalytic mechanisms. Read More View Full Article Download PDF |
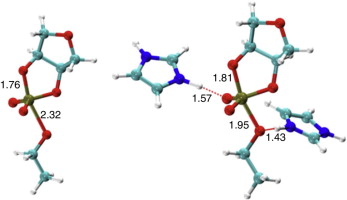 | Altered (transition) states: mechanisms of solution and enzyme catalyzed RNA 2'-O-transphosphorylation Current Opinion in Chemical Biology (2014) 21, 96-102 DOI: 10.1016/j.cbpa.2014.06.010. PMC4149925Although there have been great strides in defining the mechanisms of RNA strand cleavage by 2'-O-transphosphorylation, long-standing questions remain. How do different catalytic modes such as acid/base and metal ion catalysis influence transition state charge distribution? Does the large rate enhancement characteristic of biological catalysis result in different transition states relative to solution reactions? Answering these questions is important for understanding biological catalysis in general, and revealing principles for designing small molecule inhibitors. Recent application of linear free energy relationships and kinetic isotope effects together with multi-scale computational simulations are providing tentative answers to these questions for this fundamentally important class of phosphoryl transfer reactions. Read More View Full Article Download PDF |
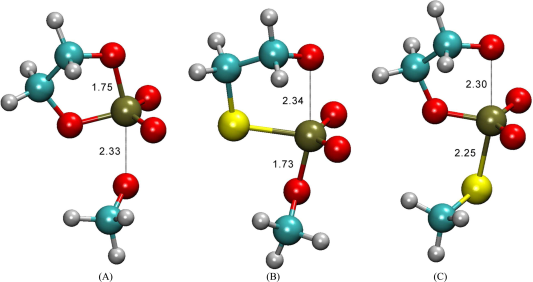 | Ab Initio Path-Integral Calculations of Kinetic and Equilibrium Isotope Effects on Base-Catalyzed RNA Transphosphorylation Models Journal of Computational Chemistry (2014) 35, 1302-1316 DOI: 10.1002/jcc.23628 PMC4096342Detailed understandings of the reaction mechanisms of RNA catalysis in various environments can have profound importance for many applications, ranging from the design of new biotechnologies to the unraveling of the evolutionary origin of life. An integral step in the nucleolytic RNA catalysis is self-cleavage of RNA strands by 2'-O-transphosphorylation. Key to elucidating a reaction mechanism is determining the molecular structure and bonding characteristics of transition state. A direct and powerful probe of transition state is measuring isotope effects on biochemical reactions, particularly if we can reproduce isotope effect values from quantum calculations. This article significantly extends the scope of our previous joint experimental and theoretical work in examining isotope effects on enzymatic and nonenzymatic 2'-O-transphosphorylation reaction models that mimic reactions catalyzed by RNA enzymes (ribozymes), and protein enzymes such as ribonuclease A (RNase A). Native reactions are studied, as well as reactions with thio substitutions representing chemical modifications often used in experiments to probe mechanism. Here, we report and compare results from eight levels of electronic-structure calculations for constructing the potential energy surfaces in kinetic and equilibrium isotope effects (KIE and EIE) computations, including a “gold-standard” coupled-cluster level of theory [CCSD(T)]. In addition to the widely used Bigeleisen equation for estimating KIE and EIE values, internuclear anharmonicity and quantum tunneling effects were also computed using our recently developed ab initio path-integral method, that is, automated integration-free path-integral method. The results of this work establish an important set of benchmarks that serve to guide calculations of KIE and EIE for RNA catalysis. Read More View Full Article Download PDF |
 | Molecular Dynamics Simulation of Nitrobenzene Dioxygenase Using AMBER Force Field Journal of Chemical Theory and Computation (2014) 10, 2246-2254 DOI: 10.1021/ct500205z PMC4059247Molecular dynamics simulation of the oxygenase component of nitrobenzene dioxygenase (NBDO) system, a member of the naphthalene family of Rieske nonheme iron dioxygenases, has been carried out using the AMBER force field combined with a new set of parameters for the description of the mononuclear nonheme iron center and iron–sulfur Rieske cluster. Simulation results provide information on the structure and dynamics of nitrobenzene dioxygenase in an aqueous environment and shed light on specific interactions that occur in its catalytic center. The results suggest that the architecture of the active site is stabilized by key hydrogen bonds, and Asn258 positions the substrate for oxidation. Analysis of protein–water interactions reveal the presence of a network of solvent molecules at the entrance to the active site, which could be of potential catalytic importance. Read More View Full Article Download PDF |
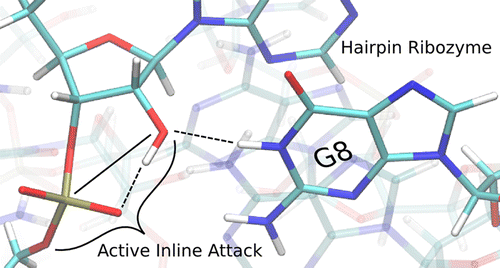 | Evidence for the role of active site residues in the hairpin ribozyme from molecular simulations along the reaction path Journal of the American Chemical Society (2014) 136, 7789-7792 DOI: 10.1021/ja500180q PMC4132952The hairpin ribozyme accelerates a phosphoryl transfer reaction without catalytic participation of divalent metal ions. Residues A38 and G8 have been implicated as playing roles in general acid and base catalysis, respectively. Here we explore the structure and dynamics of key active site residues using more than 1 µs of molecular dynamics simulations of the hairpin ribozyme at different stages along the catalytic pathway. Analysis of results indicates hydrogen bond interactions between the nucleophile and proR nonbridging oxygen are correlated with active inline attack conformations. Further, the simulation results suggest a possible alternative role for G8 to promote inline fitness and facilitate activation of the nucleophile by hydrogen bonding, although this does not necessarily exclude an additional role as a general base. Finally, we suggest that substitution of G8 with N7- or N3-deazaguanosine which have elevated pKa values, both with and without thio modifications at the 5' leaving group position, would provide valuable insight into the specific role of G8 in catalysis. Read More View Full Article Download PDF |
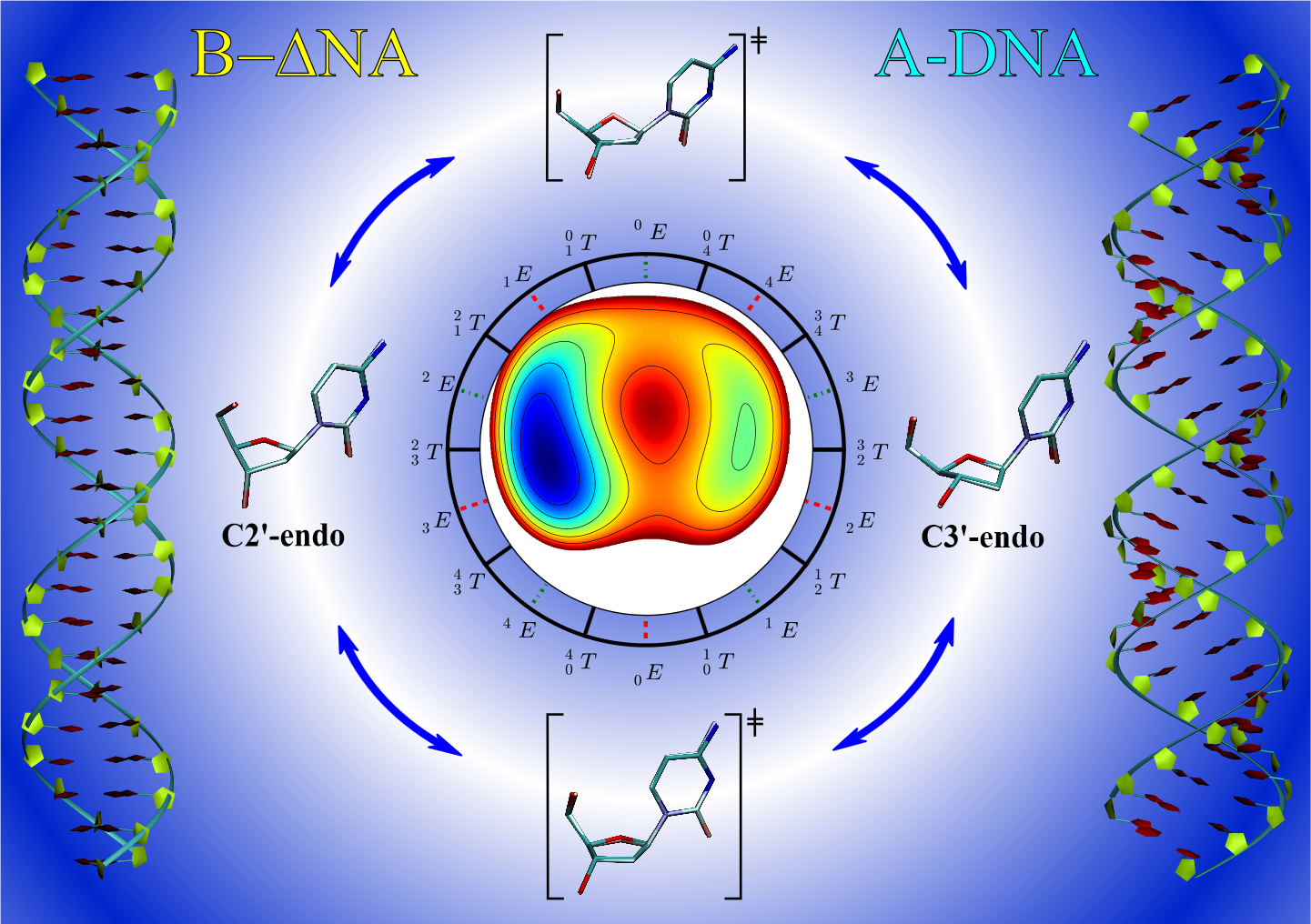 | Improvement of DNA and RNA sugar pucker profiles from semiempirical quantum methods Journal of Chemical Theory and Computation (2014) 10, 1538-1545 DOI: 10.1021/ct401013s PMC3985690Neglect of diatomic differential overlap (NDDO) and self-consistent density-functional tight-binding (SCC-DFTB) semiempirical models commonly employed in combined quantum mechanical/molecular mechanical simulations fail to adequately describe the deoxyribose and ribose sugar ring puckers. This failure limits the application of these methods to RNA and DNA systems. In this work, we provide benchmark ab initio gas-phase two-dimensional potential energy scans of the RNA and DNA sugar puckering. The benchmark calculations are compared with semiempirical models. Pucker corrections are introduced into the semiempirical models via B-spline interpolation of the potential energy difference surface relative to the benchmark data. The corrected semiempirical models are shown to well reproduce the ab initio puckering profiles. Furthermore, we demonstrate that the uncorrected semiempirical models do not usually produce a transition state between the A-form and B-form sugar puckers, but the ab initio transition state is reproduced when the B-spline correction is used. Read More View Full Article Download PDF |
 | Parametrization of an Orbital-Based Linear-Scaling Quantum Force Field for Noncovalent Interactions Journal of Chemical Theory and Computation (2014) 10, 1086-1098 DOI: 10.1021/ct401035t PMC3985928We parametrize a linear-scaling quantum mechanical force field called mDC for the accurate reproduction of nonbonded interactions. We provide a new benchmark database of accurate ab initio interactions between sulfur-containing molecules. A variety of nonbond databases are used to compare the new mDC method with other semiempirical, molecular mechanical, {\em ab initio}, and combined semiempirical quantum mechanical/molecular mechanical methods. It is shown that the molecular mechanical force field significantly and consistently reproduces the benchmark results with greater accuracy than the semiempirical models, and our mDC model produces errors twice as small as the molecular mechanical force field. The comparisons between the methods are extended to the docking of drug candidates to the Cyclin-Dependent Kinase 2 protein receptor. We correlate the protein-ligand binding energies to their experimental inhibition constants and find that the mDC produces the best correlation. Condensed phase simulation of mDC water is performed and shown to produce O-O radial distribution functions similar to TIP4P-EW Read More View Full Article Download PDF |
 | Constant pH Replica Exchange Molecular Dynamics in Explicit Solvent Using Discrete Protonation States: Implementation, Testing, and Validation Journal of Chemical Theory and Computation (2014) 10, 1341-1352 DOI: 10.1021/ct401042b PMC3985686By utilizing Graphics Processing Units, we show that constant pH molecular dynamics simulations (CpHMD) run in Generalized Born (GB) implicit solvent for long time scales can yield poor pKa predictions as a result of sampling unrealistic conformations. To address this shortcoming, we present a method for performing constant pH molecular dynamics simulations (CpHMD) in explicit solvent using a discrete protonation state model. The method involves standard molecular dynamics (MD) being propagated in explicit solvent followed by protonation state changes being attempted in GB implicit solvent at fixed intervals. Replica exchange along the pH-dimension (pH-REMD) helps to obtain acceptable titration behavior with the proposed method. We analyzed the effects of various parameters and settings on the titration behavior of CpHMD and pH-REMD in explicit solvent, including the size of the simulation unit cell and the length of the relaxation dynamics following protonation state changes. We tested the method with the amino acid model compounds, a small pentapeptide with two titratable sites, and hen egg white lysozyme (HEWL). The proposed method yields superior predicted pKa values for HEWL over hundreds of nanoseconds of simulation relative to corresponding predicted values from simulations run in implicit solvent. Read More View Full Article Download PDF |
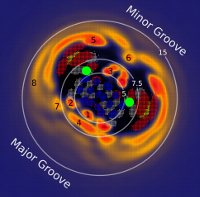 | Ion counting from explicit solvent simulations and 3D-RISM Biophysical Journal (2014) 106, 883-894 DOI: 10.1016/j.bpj.2014.01.021 PMC3944826The ionic atmosphere around nucleic acids remains only partially understood at atomic level detail in a quantitative sense. Ion counting (IC) experiments provide a quantitative measure of the ionic atmosphere around nucleic acids and, as such, are a natural route for testing quantitative theoretical approaches. In this paper we replicate IC experiments involving duplex DNA in NaCl (aq) using molecular dynamics (MD), three dimensional interaction site model (3D-RISM) and non-linear Poisson-Boltzmann (NLPB) and test against recent buffer-equilibration atomic emission spectroscopy measurements. Near physiological concentrations, MD and 3D-RISM estimates are close to experiment, but at higher concentrations (<0.7 M) both methods underestimate the number of condensed cations. NLPB calculations systematically underestimate the amount of condensed cations at almost all concentrations. The effect of DNA charge on ion and water atmosphere extends 20-25 A from its surface, yielding layered density profiles in the case of MD and 3D-RISM but less structured for NLPB. Ion distributions from 3D-RISM are relatively close to those from corresponding MD simulations, with less Na + binding in grooves and tighter binding to phosphates. We outline the statistical mechanical basis of interpreting ion counting experiments and clarify the use of specific concentration scales. Read More View Full Article Download PDF |
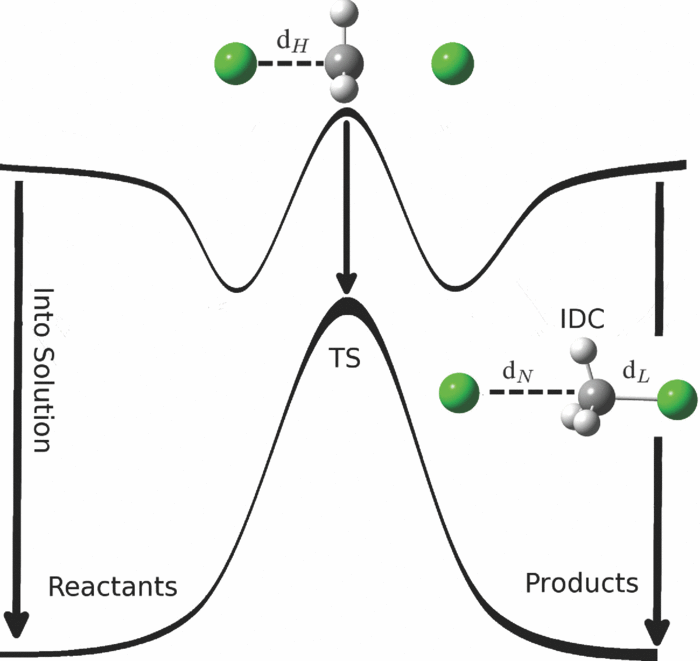 | Quantum mechanical study of solvent effects in prototype SN2 reactions in solution: Cl- attack on CH3Cl The Journal of Chemical Physics (2014) 140, 054109 DOI: 10.1063/1.4863344 PMC3977776The nucleophilic attack of a chloride ion on methyl chloride is an important prototype SN2 reaction in organic chemistry that is known to be sensitive to the effects of the surrounding solvent. Herein, we develop a highly accurate Specific Reaction Parameter (SRP) model based on the Austin Model 1 Hamiltonian for chlorine to study the effects of solvation into an aqueous environment on the reaction mechanism. To accomplish this task, we apply high-level quantum mechanical calculations to study the reaction in the gas phase and combined quantum mechanical/molecular mechanical simulations with TIP3P and TIP4P-ew water models and the resulting free energy profiles are compared with those determined from simulations using other fast semi-empirical quantum models. Both gas phase and solution results with the SRP model agree very well with experiment and provide insight into the specific role of solvent on the reaction coordinate. Overall, the newly parameterized SRP Hamiltonian is able to reproduce both the gas phase and solution phase barriers, suggesting it is an accurate and robust model for simulations in the aqueous phase at greatly reduced computational cost relative to comparably accurate ab initio and density functional models. Read More View Full Article Download PDF |
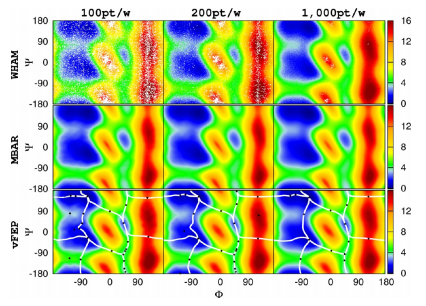 | Roadmaps through Free Energy Landscapes Calculated Using the Multidimensional vFEP Approach Journal of Chemical Theory and Computation (2014) 10, 24-34 DOI: 10.1021/ct400691f PMC3912246The variational free energy profile (vFEP) method is extended to two dimensions and tested with molecular simulation applications. The proposed 2D-vFEP approach effectively addresses the two major obstacles to constructing free energy profiles from simulation data using traditional methods: the need for overlap in the reweighting procedure and the problem of data representation. This is especially evident as these problems are shown to be more severe in two dimensions. The vFEP method is demonstrated to be highly robust and able to provide stable analytic free energy profiles with only a paucity of sampled data. The analytic profiles can be analyzed with conventional search methods to easily identify stationary points (e.g., minima and first-order saddle points) as well as the pathways that connect these points. These “roadmaps” through the free energy surface are useful not only as a post-processing tool to characterize mechanisms but can also serve as a basis from which to direct more focused “on-the-fly” sampling or adaptive force biasing. Test cases demonstrate that 2D-vFEP outperforms other methods in terms of the amount and sparsity of the data needed to construct stable, converged, analytic free energy profiles. In a classic test case, the two-dimensional free energy profile of the backbone torsion angles of alanine dipeptide, 2D-vFEP, needs less than 1% of the original data set to reach a sampling accuracy of 0.5 kcal/mol in free energy shifts between windows. A new software tool for performing one- and two-dimensional vFEP calculations is herein described and made publicly available. Read More View Full Article Download PDF |
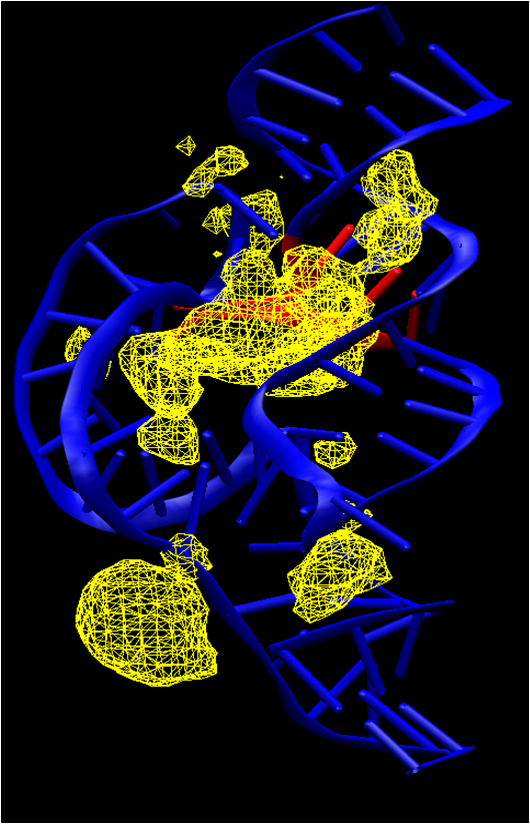 | Bridging the Gap Between Theory and Experiment to Derive a Detailed Understanding of Hammerhead Ribozyme Catalysis Progress in Molecular Biology and Translational Science (2013) 120, 25-91 DOI: 10.1016/B978-0-12-381286-5.00002-0 PMC4747252Herein we summarize our progress toward the understanding of hammerhead ribozyme (HHR) catalysis through a multiscale simulation strategy. Simulation results collectively paint a picture of HHR catalysis: HHR first folds to form an electronegative active site pocket to recruit a threshold occupation of cationic charges, either a Mg2+ ion or multiple monovalent cations. Catalytically active conformations that have good in-line fitness are supported by specific metal ion coordination patterns that involve either a bridging Mg2+ ion or multiple Na+ ions, one of which is also in a bridging coordination pattern. In the case of a single Mg2+ ion bound in the active site, the Mg2+ ion undergoes a migration that is coupled with deprotonation of the nucleophile (C17:O2'). As the reaction proceeds, the Mg2+ ion stabilizes the accumulating charge of the leaving group and significantly increases the general acid ability of G8:O2'. Further computational mutagenesis simulations suggest that the disruptions due to mutations may severely impact HHR catalysis at different stages of the reaction. Catalytic mechanisms supported by the simulation results are consistent with available structural and biochemical experiments, and together they advance our understanding of HHR catalysis. Read More View Full Article Download PDF |
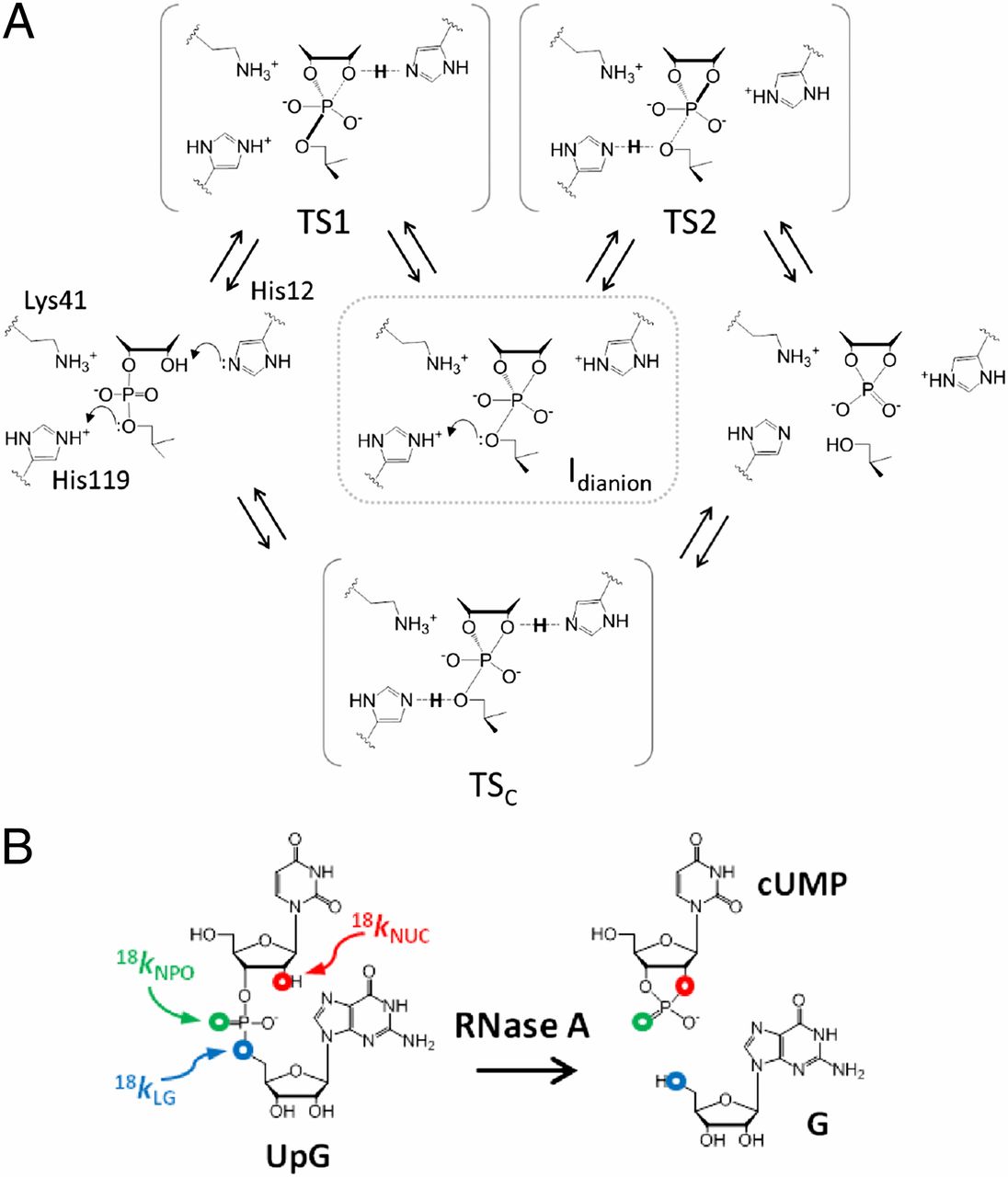 | Experimental and computational analysis of the transition state for ribonuclease A-catalyzed RNA 2'-O-transphosphorylation Proceedings of the National Academy of Sciences of the United States of America (2013) 110, 13002-13007 DOI: 10.1073/pnas.1215086110 PMC3740856Enzymes function by stabilizing reaction transition states; therefore, comparison of the transition states of enzymatic and nonenzymatic model reactions can provide insight into biological catalysis. Catalysis of RNA 2'-O-transphosphorylation by ribonuclease A is proposed to involve electrostatic stabilization and acid/base catalysis, although the structure of the rate-limiting transition state is uncertain. Here, we describe coordinated kinetic isotope effect (KIE) analyses, molecular dynamics simulations, and quantum mechanical calculations to model the transition state and mechanism of RNase A. Comparison of the 18O KIEs on the 2'O nucleophile, 5'O leaving group, and nonbridging phosphoryl oxygens for RNase A to values observed for hydronium- or hydroxide-catalyzed reactions indicate a late anionic transition state. Molecular dynamics simulations using an anionic phosphorane transition state mimic suggest that H-bonding by protonated His12 and Lys41 stabilizes the transition state by neutralizing the negative charge on the nonbridging phosphoryl oxygens. Quantum mechanical calculations consistent with the experimental KIEs indicate that expulsion of the 5'O remains an integral feature of the rate-limiting step both on and off the enzyme. Electrostatic interactions with positively charged amino acid site chains (His12/Lys41), together with proton transfer from His119, render departure of the 5'O less advanced compared with the solution reaction and stabilize charge buildup in the transition state. The ability to obtain a chemically detailed description of 2'-O-transphosphorylation transition states provides an opportunity to advance our understanding of biological catalysis significantly by determining how the catalytic modes and active site environments of phosphoryl transferases influence transition state structure. Read More View Full Article Download PDF |
 | A Framework for Flexible and Scalable Replica-Exchange on Production Distributed Cyberinfrastructure. Proceedings of the Conference on Extreme Science and Engineering Discovery Environment: Gateway to Discovery(2013) DOI: 10.1145/2484762.2484830 Replica-Exchange represent a powerful class of algorithms that are used for enhanced configurational and energetic sampling in a range of physical systems. Computationally they represent a type of applications with multiple scales of communication; at a fine-grained level there is often communication with a replica, typically an MPI process. At a coarse-grained level -- both temporally as well as in data amount exchanged, the replicas communicate with other replicas. In this paper, we outline a novel framework that we have developed that supports the large-scale and flexible execution of a number of replicas. The framework is flexible in the sense that it supports different coupling schemes between the replicas, and is agnostic to the specific underlying simulation -- classical or quantum, single-core simulation or a parallel simulation. As a measure of the scalability of the framework, we measure the number of nanoseconds simulated a day, as a function of the number of replicas. In spite of the increasing communication and coordination requirements as a function of the number of replicas, our framework supports the execution of a thousand replicas without significant overhead. Furthermore, as representative of the efficiency of the framework, the number of nanoseconds simulated in twenty-four hours as a function of replicas remains essentially constant. Although there are several specific aspects that will benefit from further optimization, a first working prototype has the ability to fundamentally change the scale of replica-exchange simulations possible on production distributed cyberinfrastructure such as XSEDE, as well as support novel usage modes on these infrastructure. This paper also represents the release of the framework to the broader biophysical simulation community and provides details on its usage. Read More View Full Article Download PDF |
 | A Variational Linear-Scaling Framework to Build Practical, Efficient Next-Generation Orbital-Based Quantum Force Fields Journal of Chemical Theory and Computation (2013) 9, 1417-1427 DOI: 10.1021/ct3010134 PMC3694615We introduce a new hybrid molecular orbital/density-functional modified divide-and-conquer (mDC) approach that allows the linear-scaling calculation of very large quantum systems. The method provides a powerful framework from which linear-scaling force fields for molecular simulations can be developed. The method is variational in the energy and has simple, analytic gradients and essentially no break-even point with respect to the corresponding full electronic structure calculation. Furthermore, the new approach allows intermolecular forces to be properly balanced such that nonbonded interactions can be treated, in some cases, to much higher accuracy than the full calculation. The approach is illustrated using the second-order self-consistent charge density-functional tight-binding model (DFTB2). Using this model as a base Hamiltonian, the new mDC approach is applied to a series of water systems, where results show that geometries and interaction energies between water molecules are greatly improved relative to full DFTB2. In order to achieve substantial improvement in the accuracy of intermolecular binding energies and hydrogen bonded cluster geometries, it was necessary to extend the DFTB2 model to higher-order atom-centered multipoles for the second-order self-consistent intermolecular electrostatic term. Using generalized, linear-scaling electrostatic methods, timings demonstrate that the method is able to calculate a water system of 3000 atoms in less than half of a second, and systems of up to 1 million atoms in only a few minutes using a conventional desktop workstation. Read More View Full Article Download PDF |
 | Molecular Simulations of RNA 2'-O-Transesterification Reaction Models in Solution The Journal of Physical Chemistry B (2013) 117, 94-103 DOI: 10.1021/jp3084277 PMC3574632We employ quantum mechanical/molecular mechanical umbrella sampling simulations to probe the free energy surfaces of a series of increasingly complex reaction models of RNA 2'-O-transesterification in aqueous solution under alkaline conditions. Such models are valuable for understanding the uncatalyzed processes underlying catalytic cleavage of the phosphodiester backbone of RNA, a reaction of fundamental importance in biology. The chemically reactive atoms are modeled by the AM1/d-PhoT quantum model for phosphoryl transfer, whereas the aqueous solvation environment is modeled with a molecular mechanics force field. Several simulation protocols were compared that used different ionic conditions and force field models. The results provide insight into how variation of the structural environment of the nucleophile and leaving group affects the free energy profile for the transesterification reaction. Results for a simple RNA backbone model are compared with recent experiments by Harris et al. on the specific base-catalyzed cleavage of a UpG dinucleotide. The calculated and measured free energies of activation match extremely well (?F = 19.9–20.8 vs 19.9 kcal/mol). Solvation is seen to play a crucial role and is characterized by a network of hydrogen bonds that envelopes the pentacoordinate dianionic phosphorane transition state and provides preferential stabilization relative to the reactant state. Read More View Full Article Download PDF |
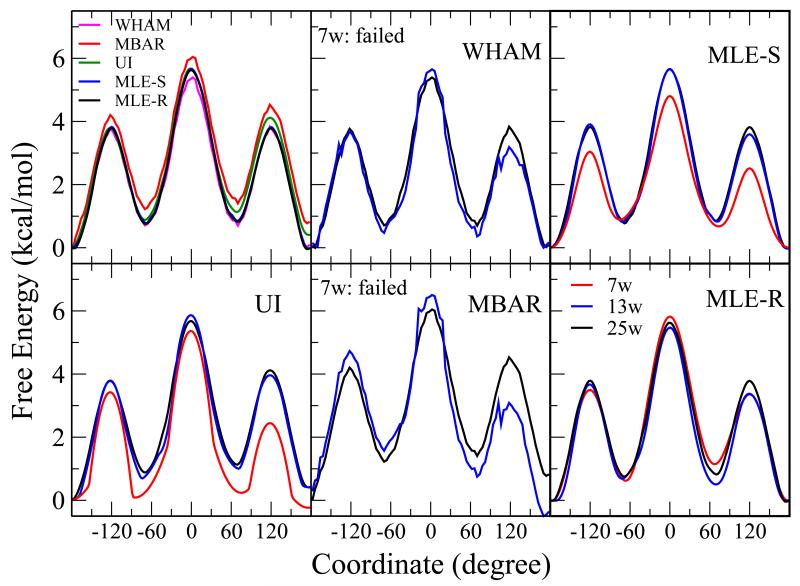 | A New Maximum Likelihood Approach for Free Energy Profile Construction from Molecular Simulations Journal of Chemical Theory and Computation (2013) 9, 153-164 DOI: 10.1021/ct300703z PMC3580863A novel variational method for construction of free energy profiles from molecular simulation data is presented. The variational free energy profile (VFEP) method uses the maximum likelihood principle applied to the global free energy profile based on the entire set of simulation data (e.g., from multiple biased simulations) that spans the free energy surface. The new method addresses common obstacles in two major problems usually observed in traditional methods for estimating free energy surfaces: the need for overlap in the reweighting procedure and the problem of data representation. Test cases demonstrate that VFEP outperforms other methods in terms of the amount and sparsity of the data needed to construct the overall free energy profiles. For typical chemical reactions, only 5 windows and 20–35 independent data points per window are sufficient to obtain an overall qualitatively correct free energy profile with sampling errors an order of magnitude smaller than the free energy barrier. The proposed approach thus provides a feasible mechanism to quickly construct the global free energy profile and identify free energy barriers and basins in free energy simulations via a robust, variational procedure that determines an analytic representation of the free energy profile without the requirement of numerically unstable histograms or binning procedures. It can serve as a new framework for biased simulations and is suitable to be used together with other methods to tackle the free energy estimation problem. Read More View Full Article Download PDF |
 | Exact Relation between Potential of Mean Force and Free-Energy Profile Journal of Chemical Theory and Computation (2012) 8, 3998-4003 DOI: 10.1021/ct300392f PMC3505112We apply concepts of covariant and contravariant vector space in differential geometry and general relativity to derive new, general, exact relations between potential of mean force and free-energy profile. These relations are immensely practical in free-energy simulations because a full Jacobian transformation (which is usually unknown) is not required; rather, only knowledge of the (constraint) coordinate of interest is needed. We reveal that in addition to the Jacobian determinant, the Jacobian scale factor and Leibnizian contributions must also be considered, as well as a Fixman term with correct mass dependence. Our newly derived relations are verified with new nontrivial benchmark numerical examples for which exact results can be computed and compared with relations available in the literature that turn out to exhibit significant deviations from the exact values. Read More View Full Article Download PDF |
 | Mapping L1 ligase ribozyme conformational switch Journal of Molecular Biology (2012) 423, 106-122 DOI: 10.1016/j.jmb.2012.06.035 PMC3509952 L1 ligase (L1L) molecular switch is an in vitro optimized synthetic allosteric ribozyme that catalyzes the regioselective formation of a 5'-to-3' phosphodiester bond, a reaction for which there is no known naturally occurring RNA catalyst. L1L serves as a proof of principle that RNA can catalyze a critical reaction for prebiotic RNA self-replication according to the RNA world hypothesis. L1L crystal structure captures two distinct conformations that differ by a reorientation of one of the stems by around 80 Å and are presumed to correspond to the active and inactive state, respectively. It is of great interest to understand the nature of these two states in solution and the pathway for their interconversion. In this study, we use explicit solvent molecular simulation together with a novel enhanced sampling method that utilizes concepts from network theory to map out the conformational transition between active and inactive states of L1L. We find that the overall switching mechanism can be described as a three-state/two-step process. The first step involves a large-amplitude swing that reorients stem C. The second step involves the allosteric activation of the catalytic site through distant contacts with stem C. Using a conformational space network representation of the L1L switch transition, it is shown that the connection between the three states follows different topographical patterns: the stem C swing step passes through a narrow region of the conformational space network, whereas the allosteric activation step covers a much wider region and a more diverse set of pathways through the network. Read More View Full Article Download PDF |
 | Extended Polarization in Third-Order SCC-DFTB from Chemical-Potential Equalization The Journal of Physical Chemistry A (2012) 116, 9131-9141 DOI: 10.1021/jp306239c PMC3546173In this work, we augment the approximate density functional method SCC-DFTB (DFTB3) with the chemical-potential equalization (CPE) approach in order to improve the performance for molecular electronic polarizabilities. The CPE method, originally implemented for the NDDO type of methods by Giese and York, has been shown to significantly emend minimal basis methods with respect to the response properties and has been applied to SCC-DFTB recently. CPE allows this inherent limitation of minimal basis methods to be overcome by supplying an additional response density. The systematic underestimation is thereby corrected quantitatively without the need to extend the atomic orbital basis (i.e., without increasing the overall computational cost significantly). The dependency of polarizability as a function of the molecular charge state, especially, was significantly improved from the CPE extension of DFTB3. The empirical parameters introduced by the CPE approach were optimized for 172 organic molecules in order to match the results from density functional theory methods using large basis sets. However, the first-order derivatives of molecular polarizabilities (e.g., required to compute Raman activities) are not improved by the current CPE implementation (i.e., Raman spectra are not improved). Read More View Full Article Download PDF |
 | Density-functional expansion methods: Grand challenges Theoretical Chemistry Accounts (2012) 131, 1145-1161 DOI: 10.1007/s00214-012-1145-7 PMC4898065We discuss the source of errors in semiempirical density-functional expansion (VE) methods. In particular, we show that VE methods are capable of well reproducing their standard Kohn-Sham density-functional method counterparts, but suffer from large errors upon using one or more of these approximations: the limited size of the atomic orbital basis, the Slater monopole auxiliary basis description of the response density, and the one- and two-body treatment of the core-Hamiltonian matrix elements. In the process of discussing these approximations and highlighting their symptoms, we introduce a new model that supplements the second-order density-functional tight-binding model with a self-consistent charge-dependent chemical potential equalization correction; we review our recently reported method for generalizing the auxiliary basis description of the atomic orbital response density; and we decompose the first-order potential into a summation of additive atomic components and many-body corrections, and from this examination, we provide new insights and preliminary results that motivate and inspire new approximate treatments of the core-Hamiltonian. Read More View Full Article Download PDF |
 | Characterization of the Reaction Path and Transition States for RNA Transphosphorylation Models from Theory and Experiment Angewandte Chemie International Edition (2012) 51, 647-651 DOI: 10.1002/anie.201104147 PMC3448066The primary and secondary kinetic isotope effects for a model compound which represents RNA cleavage transesterification were calculated and compared with experimental measurements. Based on the good agreement between theory and experiments, the energy profile, reaction pathway, and two distinct transition states for the reactions of the model compound and two thio-substituted analogues were characterized. Read More View Full Article Download PDF |
 | Characterization of the structure and dynamics of the HDV ribozyme in different stages along the reaction path The Journal of Physical Chemistry Letters (2011) 2, 2538-2543 DOI: 10.1021/jz201106y PMC3244300The structure and dynamics of the hepatitis delta virus ribozyme (HDVr) are studied using molecular dynamics simulations in several stages along its catalytic reaction path, including reactant, activated precursor, and transition-state mimic and product states, departing from an initial structure based on the C75U mutant crystal structure (PDB: 1VC7). Results of five 350 ns molecular dynamics simulations reveal a spontaneous rotation of U-1 that leads to an in-line conformation and supports the role of protonated C75 as the general acid in the transition state. Our results provide rationale for the interpretation of several important experimental results and make experimentally testable predictions regarding the roles of key active site residues that are not obvious from any available crystal structures. Read More View Full Article Download PDF |
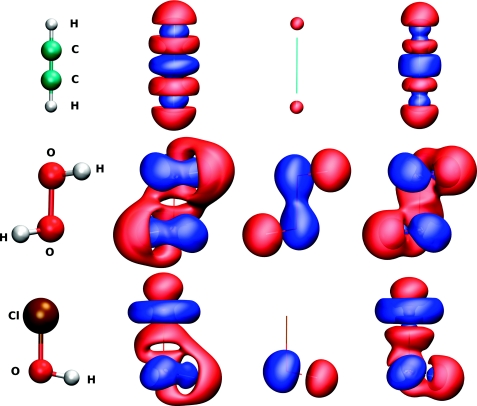 | Density-functional expansion methods: Generalization of the auxiliary basis The Journal of Chemical Physics (2011) 134, 194103 DOI: 10.1063/1.3587052 PMC3113327The formulation of density-functional expansion methods is extended to treat the second and higher-order terms involving the response density and spin densities with an arbitrary single-center auxiliary basis. The two-center atomic orbital products are represented by the auxiliary functions centered about those two atoms, and the mapping coefficients are determined from a local constrained variational procedure. This two-center variational procedure allows the mapping coefficients to be pretabulated and splined as a function of internuclear separation for efficient look up. The splines of mapping coefficients have a range no longer than that of the overlap integrals, and the auxiliary density appears as a single point-multipole expansion to all nonoverlapping atoms, thus allowing for the trivial implementation of a linear-scaling algorithm. The method is tested using Gaussian multipole expansions, and the effect of angular and radial completeness is explored. Several auxiliary basis sets are parametrized and compared to an auxiliary basis analogous to that used in the self-consistent-charge density-functional tight-binding model, and the method is demonstrated to greatly improve the representation of the density response with respect to a reference expansion model that does not use an auxiliary basis. Read More View Full Article Download PDF |
 | Influence of C-5 substituted cytosine and related nucleoside analogs on the formation of benzo[a]pyrene diol epoxide-dG adducts Nucleic Acids Research (2011) 39, 3988-4006 DOI: 10.1093/nar/gkq1341 PMC3089471Endogenous 5-methylcytosine (MeC) residues are found at all CG dinucleotides of the p53 tumor suppressor gene, including the mutational ‘hotspots’ for smoking induced lung cancer. MeC enhances the reactivity of its base paired guanine towards carcinogenic diolepoxide metabolites of polycyclic aromatic hydrocarbons (PAH) present in cigarette smoke. In the present study, the structural basis for these effects was investigated using a series of unnatural nucleoside analogs and a representative PAH diolepoxide, benzo[a]pyrene diolepoxide (BPDE). Synthetic DNA duplexes derived from a frequently mutated region of the p53 gene (5'-CCCGGCACCC GC[15N3,13C1-G]TCCGCG-3', + strand) were prepared containing [15N3, 13C1]-guanine opposite unsubstituted cytosine, MeC, abasic site, or unnatural nucleobase analogs. Following BPDE treatment and hydrolysis of the modified DNA to 2'-deoxynucleosides, N2-BPDE-dG adducts formed at the [15N3, 13C1]-labeled guanine and elsewhere in the sequence were quantified by mass spectrometry. We found that C-5 alkylcytosines and related structural analogs specifically enhance the reactivity of the base paired guanine towards BPDE and modify the diastereomeric composition of N2-BPDE-dG adducts. Fluorescence and molecular docking studies revealed that 5-alkylcytosines and unnatural nucleobase analogs with extended aromatic systems facilitate the formation of intercalative BPDE–DNA complexes, placing BPDE in a favorable orientation for nucleophilic attack by the N2 position of guanine. Read More View Full Article Download PDF |
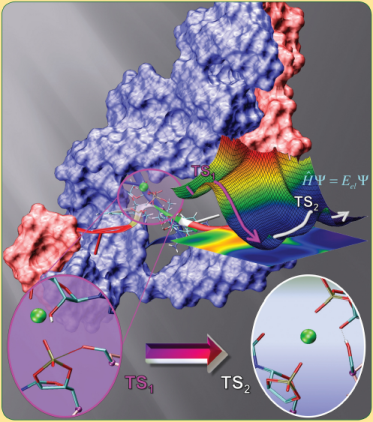 | Active participation of Mg2+ ion in the reaction coordinate of RNA self-cleavage catalyzed by the hammerhead ribozymes Journal of Chemical Theory and Computation (2011) 7, 1-3 DOI: 10.1021/ct100467t PMC3046872We report results from combined quantum mechanical/molecular mechanical (QM/MM) free energy simulations to explore metal-assisted phosphoryl transfer and general acid catalysis in the extended hammerhead ribozyme. The mechanisms considered here assume that the 2'OH group of C17 has already been activated (i.e., is deprotonated) and acts as a nucleophile to go on an in-line attack to the adjacent scissile phosphate, passing through a pentavalent phosphorane intermediate/transition state, followed by acid-catalyzed departure of the O5' leaving group of C1.1. A series of six two-dimensional potential of mean force profiles are reported in this study, requiring an aggregate of over 100 ns of QM/MM simulation. The simulations employ the AM1/d-PhoT semiempirical quantum model and linear-scaling QM/MM-Ewald method and explore mechanistic pathways for the self-cleavage. Results support the plausibility of a cleavage mechanism where phosphoryl transfer and general acid catalysis are stepwise, and where the catalytic divalent metal ion plays an active role in the chemical steps of catalysis. Read More View Full Article Download PDF |
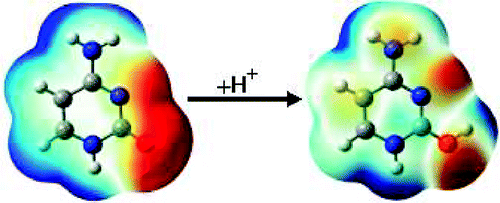 | Accurate proton affinity and gas-phase basicity values of molecules important in biocatalysis The Journal of Physical Chemistry B (2010) 114, 13911-13921 DOI: 10.1021/jp107450n PMC2970571Benchmark quantum calculations of proton affinities and gas-phase basicities of molecules relevant to biochemical processes, particularly acid/base catalysis, are presented and compared for a variety of multilevel and density functional quantum models. Included are nucleic acid bases in both keto and enol tautomeric forms, ribose in B-form and A-form sugar pucker conformations, amino acid side chains and backbone molecules, and various phosphates and phosphoranes, including thio substitutions. This work presents a high-level thermodynamic characterization of biologically relevant protonation states and provides a benchmark database for development of next-generation semiempirical and approximate density functional quantum models and parametrization of methods to predict pKa values and relative solvation energies. Read More View Full Article Download PDF |
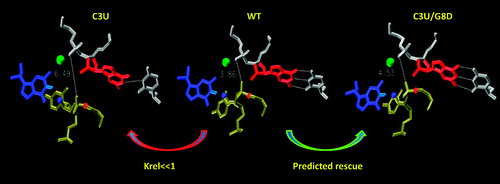 | Computational mutagenesis studies of hammerhead ribozyme catalysis Journal of the American Chemical Society (2010) 132, 13505-13518 DOI: 10.1021/ja105956u PMC2948978Computational studies of the mutational effects at the C3, G8, and G5 positions of the hammerhead ribozyme (HHR) are reported, based on a series of twenty-four 100-ns molecular dynamics simulations of the native and mutated HHR in the reactant state and in an activated precursor state (G8:2'OH deprotonated). Invoking the assumptions that G12 acts as the general base while the 2'OH of G8 acts as a general acid, the simulations are able to explain the origins of experimentally observed mutational effects, including several that are not easily inferred from the crystal structure. Simulations suggest that the Watson-Crick base-pairing between G8 and C3, the hydrogen bond network between C17 and G5, and the base stacking interactions between G8 and C1.1, collectively, are key to maintaining an active site structure conducive for catalytic activity. Mutation-induced disruption of any of these interactions will adversely affect activity. The simulation results predict that the C3U/G8D double mutant, where D is 2,6-diaminopurine, will have a rescue effect relative to the corresponding single mutations. Two general conclusions about the simulations emerge from this work. First, mutation simulations may require 30 ns or more to suitably relax such that the mutational effects become apparent. Second, in some cases, it is necessary to look beyond the reactant state in order to interpret mutational effects in terms of catalytically active structure. The present simulation results lead to better understanding of the origin of experimental mutational effects and provide insight into the key conserved features necessary to maintain the integrity of the active site architecture. Read More View Full Article Download PDF |
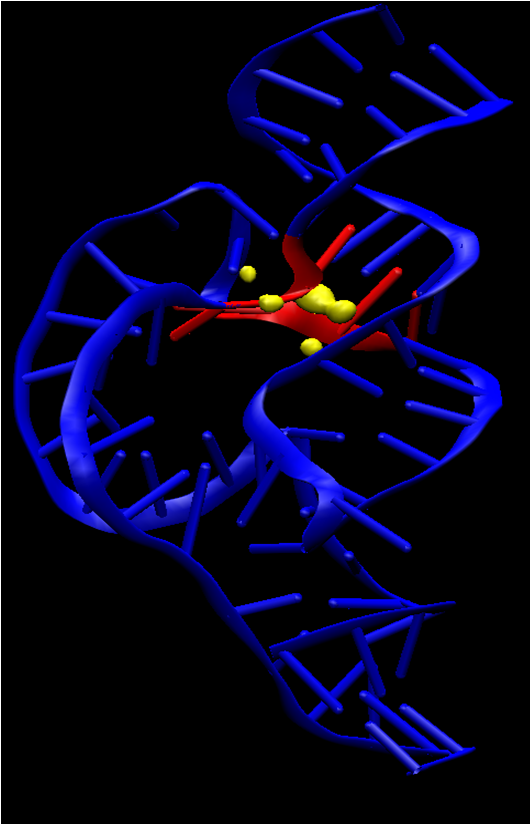 | Insights into the Role of Conformational Transitions and Metal Ion Binding in RNA Catalysis from Molecular Simulations In Annual Reports in Computational Chemistry Annual Reports in Computational Chemistry, Ralph A. Wheeler ed.(2010) 6, 168-200 DOI: 10.1016/S1574-1400(10)06001-9 ISBN: 978-0-44-453552-8We present a summary of recent advances in the application of molecular simulation methods to study the mechanisms of RNA catalysis. The focus of this chapter is on the nature of conformational transitions and metal ion binding on structure and activity. Two RNA enzyme systems are considered: the hammerhead ribozyme and the L1 ligase. The hammerhead ribozyme is a small archetype ribozyme that undergoes a conformational transition into a catalytically active conformation in a step that is concerted with changes in metal ion binding in the active site. The L1 ligase ribozyme is an in vitro selected ribozyme that uses a noncanonically base-paired ligation site to catalyze regioselectively and regiospecifically the 5' to 3' phosphodiester bond ligation. The L1 ligase presumably undergoes a large-scale conformational change from an inactive to an active form that involves reorientation of one of the stems by around 80 Å, making it a novel catalytic riboswitch. Analysis of the simulation results and comparison with experimental measurements provide important new insight into the conformational and chemical steps of catalysis of the hammerhead and L1 ligase ribozymes. Read More View Full Article Download PDF |
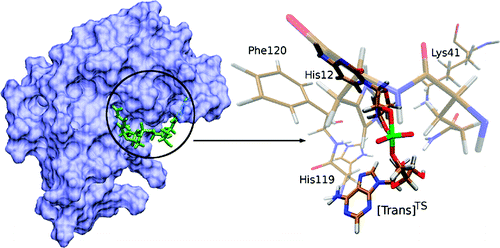 | Molecular dynamics simulation of bovine pancreatic ribonuclease A - CpA and transition state-like complexes The Journal of Physical Chemistry B (2010) 114, 7371-7382 DOI: 10.1021/jp909004y PMC2892782The mechanisms of enzymes are intimately connected with their overall structure and dynamics in solution. Experimentally, it is considerably challenging to provide detailed atomic level information about the conformational events that occur at different stages along the chemical reaction path. Here, theoretical tools may offer new potential insights that complement those obtained from experiments that may not yield an unambiguous mechanistic interpretation. In this study, we apply molecular dynamics simulations of bovine pancreatic ribonuclease A, an archetype ribonuclease, to study the conformational dynamics, structural relaxation, and differential solvation that occur at discrete stages of the transesterification and cleavage reaction. Simulations were performed with explicit solvation with rigorous electrostatics and utilize recently developed molecular mechanical force field parameters for transphosphorylation and hydrolysis transition state analogues. Herein, we present results for the enzyme complexed with the dinucleotide substrate cytidilyl-3',5'-adenosine (CpA) in the reactant, and transphosphorylation and hydrolysis transition states. A detailed analysis of active site structures and hydrogen-bond patterns is presented and compared. The integrity of the overall backbone structure is preserved in the simulations and supports a mechanism whereby His12 stabilizes accumulating negative charge at the transition states through hydrogen-bond donation to the nonbridge oxygens. Lys41 is shown to be highly versatile along the reaction coordinate and can aid in the stabilization of the dianionic transition state, while being poised to act as a general acid catalyst in the hydrolysis step. Read More View Full Article Download PDF |
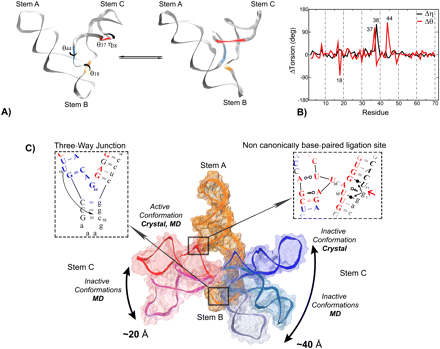 | Identification of dynamical hinge points of the L1 ligase molecular switch RNA (2010) 16, 769-780 DOI: 10.1261/rna.1897810 PMC2844624The L1 ligase is an in vitro selected ribozyme that uses a noncanonically base-paired ligation site to catalyze regioselectively and regiospecifically the 5' to 3' phosphodiester bond ligation, a reaction relevant to origin of life hypotheses that invoke an RNA world scenario. The L1 ligase crystal structure revealed two different conformational states that were proposed to represent the active and inactive forms. It remains an open question as to what degree these two conformers persist as stable conformational intermediates in solution, and along what pathway are they able to interconvert. To explore these questions, we have performed a series of molecular dynamics simulations in explicit solvent of the inactive–active conformational switch in L1 ligase. Four simulations were performed departing from both conformers in both the reactant and product states, in addition to a simulation where local unfolding in the active state was induced. From these simulations, along with crystallographic data, a set of four virtual torsion angles that span two evolutionarily conserved and restricted regions were identified as dynamical hinge points in the conformational switch transition. The ligation site visits three distinct states characterized by hydrogen bond patterns that are correlated with the formation of specific contacts that may promote catalysis. The insights gained from these simulations contribute to a more detailed understanding of the coupled catalytic/conformational switch mechanism of L1 ligase that may facilitate the design and engineering of new catalytic riboswitches. Read More View Full Article Download PDF |
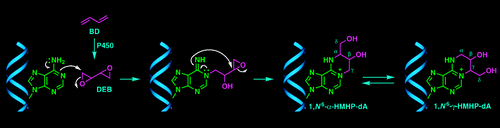 | Exocyclic deoxyadenosine adducts of 1,2,3,4-Diepoxybutane: Synthesis, structural elucidation, and mechanistic studies Chemical Research in Toxicology (2010) 23, 118-133 DOI: 10.1021/tx900312e PMC2807914 1,2,3,4-Diepoxybutane (DEB) is considered the ultimate carcinogenic metabolite of 1,3-butadiene, an important industrial chemical and environmental pollutant present in urban air. Although it preferentially modifies guanine within DNA, DEB induces a large number of A ? T transversions, suggesting that it forms strongly mispairing lesions at adenine nucleobases. We now report the discovery of three potentially mispairing exocyclic adenine lesions of DEB: N6,N6-(2,3-dihydroxybutan-1,4-diyl)-2'-deoxyadenosine (compound 2), 1,N6-(2-hydroxy-3-hydroxymethylpropan-1,3-diyl)-2'-deoxyadenosine (compound 3), and 1,N6-(1-hydroxymethyl-2-hydroxypropan-1,3-diyl)-2'-deoxyadenosine (compound 4). The structures and stereochemistry of the novel DEB-dA adducts were determined by a combination of UV and NMR spectroscopy, tandem mass spectrometry, and independent synthesis. We found that synthetic N6-(2-hydroxy-3,4-epoxybut-1-yl)-2'-deoxyadenosine (compound 1) representing the product of N6-adenine alkylation by DEB spontaneously cyclizes to form 3 under aqueous conditions or 2 under anhydrous conditions in the presence of an organic base. Compound 3 can be interconverted with 4 by a reversible unimolecular pericyclic reaction favoring 4 as a more thermodynamically stable product. Both 3 and 4 are present in double stranded DNA treated with DEB in vitro and in liver DNA of laboratory mice exposed to 1,3-butadiene by inhalation. We propose that in DNA under physiological conditions, DEB alkylates the N-1 position of adenine in DNA to form N1-(2-hydroxy-3,4-epoxybut-1-yl)-adenine adducts, which undergo an SN2-type intramolecular nucleophilic substitution and rearrangement to give 3 (minor) and 4 (major). Formation of exocyclic DEB-adenine lesions following exposure to 1,3-butadiene provides a possible mechanism of mutagenesis at the A:T base pairs. Read More View Full Article Download PDF |
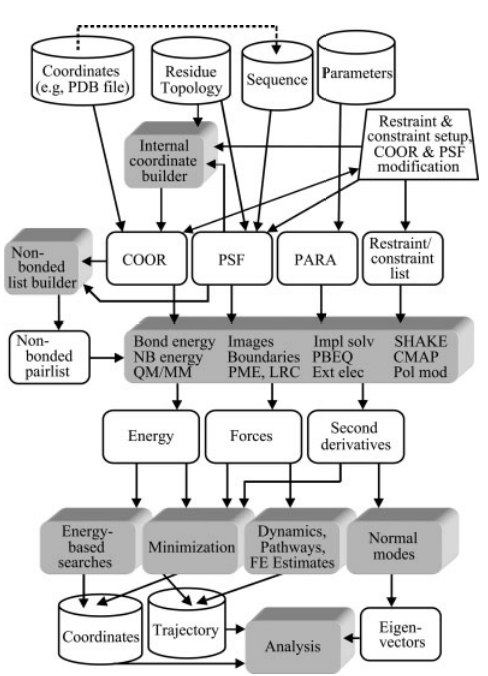 | CHARMM: The biomolecular simulation program Journal of Computational Chemistry (2009) 30, 1545-1614 DOI: 10.1002/jcc.21287 CHARMM (Chemistry at HARvard Molecular Mechanics) is a highly versatile and widely used molecular simulation program. It has been developed over the last three decades with a primary focus on molecules of biological interest, including proteins, peptides, lipids, nucleic acids, carbohydrates, and small molecule ligands, as they occur in solution, crystals, and membrane environments. For the study of such systems, the program provides a large suite of computational tools that include numerous conformational and path sampling methods, free energy estimators, molecular minimization, dynamics, and analysis techniques, and model-building capabilities. The CHARMM program is applicable to problems involving a much broader class of many-particle systems. Calculations with CHARMM can be performed using a number of different energy functions and models, from mixed quantum mechanical-molecular mechanical force fields, to all-atom classical potential energy functions with explicit solvent and various boundary conditions, to implicit solvent and membrane models. The program has been ported to numerous platforms in both serial and parallel architectures. This article provides an overview of the program as it exists today with an emphasis on developments since the publication of the original CHARMM article in 1983. © 2009 Wiley Periodicals, Inc.J Comput Chem, 2009. Read More View Full Article Download PDF |
 | Unraveling the mechanisms of ribozyme catalysis with multi-scale simulations Multi-scale Quantum Models for Biocatalysis(2009) 7, 377-408 Chapter: Multi-scale Quantum Models for Biocatalysis DOI: 10.1007/978-1-4020-9956-4_14 ISBN: 978-1-4020-9956-4Description of a multiscale simulation strategy we have developed to attack problems of RNA catalysis is presented. Ribozyme systems give special challenges not present in typical protein systems, and consequently demand new methods. The main methodological components are herein summarized, including the assembly of the QCRNA database, parameterization of the AM1/d-PhoT Hamiltonian, and development of new semiempirical functional forms for improved charge-dependent response properties, methods for coupling many-body exchange, correlation and dispersion into the QM/MM interaction, and generalized methods for linear-scaling electrostatics, solvation and solvent boundary potentials. Results for a series of case studies ranging from noncatalytic reaction models that compare the effect of new DFT functionals, and on catalytic RNA systems including the hairpin, hammerhead and L1 ligase ribozymes are discussed. Read More View Full Article Download PDF |
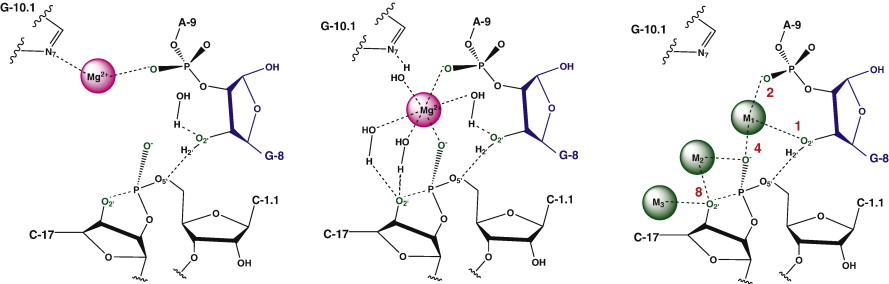 | Threshold occupancy and specific cation binding modes in the hammerhead ribozyme active site are required for active conformation Journal of Molecular Biology (2009) 388, 195-206 DOI: 10.1016/j.jmb.2009.02.054 PMC2715853The relationship between formation of active in-line attack conformations and monovalent (Na+) and divalent (Mg2+) metal ion binding in hammerhead ribozyme (HHR) has been explored with molecular dynamics simulations. To stabilize repulsions between negatively charged groups, different requirements of the threshold occupancy of metal ions were observed in the reactant and activated precursor states both in the presence and in the absence of a Mg2+ in the active site. Specific bridging coordination patterns of the ions are correlated with the formation of active in-line attack conformations and can be accommodated in both cases. Furthermore, simulation results suggest that the HHR folds to form an electronegative recruiting pocket that attracts high local concentrations of positive charge. The present simulations help to reconcile experiments that probe the metal ion sensitivity of HHR catalysis and support the supposition that Mg2+, in addition to stabilizing active conformations, plays a specific chemical role in catalysis. Read More View Full Article Download PDF |
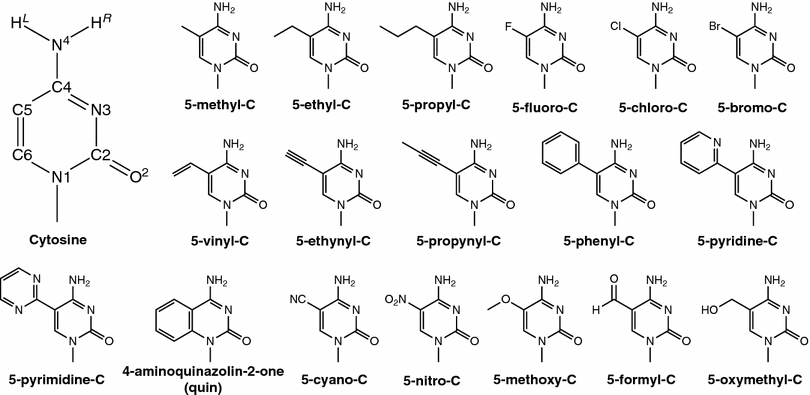 | Density functional study of the influence of C5 cytosine substitution in base pairs with guanine Theoretical Chemistry Accounts (2009) 122, 179-188 DOI: 10.1007/s00214-008-0497-5 PMC2771868 The present study employs density-functional electronic structure methods to investigate the effect of chemical modification at the C5 position of cytosine. A series of experimentally motivated chemical modifications are considered, including alkyl, halogen, aromatic, fused ring, and strong s and p withdrawing functional groups. The effect of these modifications on cytosine geometry, electronic structure, proton affinities, gas phase basicities, cytosine–guanine base pair hydrogen bond network and corresponding nucleophilicity at guanine are examined. Ultimately, these results play a part in dissecting the effect of endogenous cytosine methylation on the reactivity of neighboring guanine toward carcinogens and DNA alkylating agents. Read More View Full Article Download PDF |
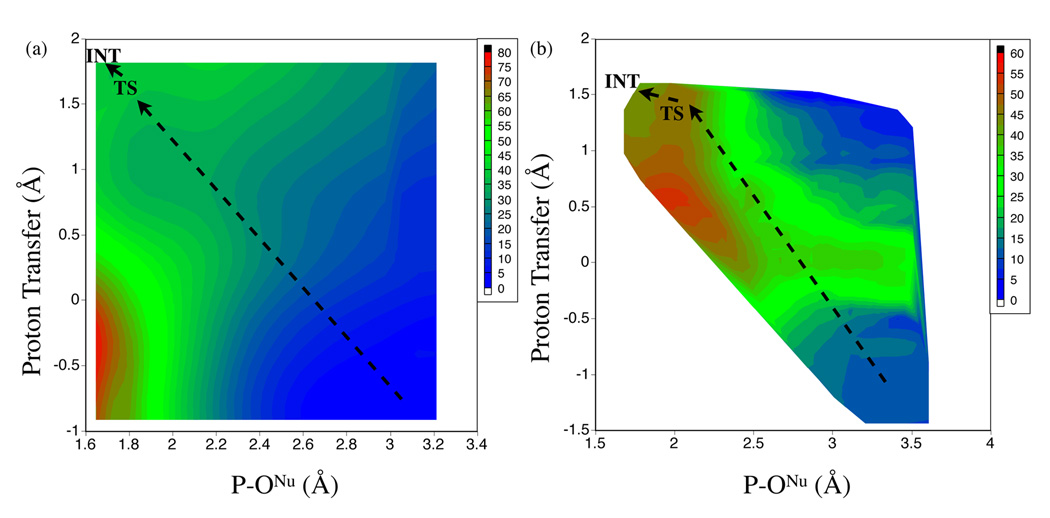 | Description of phosphate hydrolysis reactions with the self-consistent-charge density-functional-tight-binding (SCC-DFTB) theory. 1. Parameterization Journal of Chemical Theory and Computation (2008) 4, 2067-2084 DOI: 10.1021/ct800330d PMC2665970Phosphate chemistry is involved in many key biological processes yet the underlying mechanism often remains unclear. For theoretical analysis to effectively complement experimental mechanistic analysis, it is essential to develop computational methods that can capture the complexity of the underlying potential energy surface and allow for sufficient sampling of the configurational space. To this end, we report the parameterization of an approximate density functional theory, Self-Consistent-Charge Density-Functional Tight-Binding (SCC-DFTB) method for systems containing phosphorus. Compared to high-level density functional theory and ab initio (MP2 and G3B3) results, the standard second-order parameterization is shown to give reliable structures for a diverse set of phosphate compounds but inaccurate energetics. With the on-site third-order terms included, referred to as SCC-DFTBPA, calculated proton affinities of phosphate compounds are substantially improved, although it remains difficult to obtain reliable proton affinity for both phosphates and compounds that do not contain phosphorus, indicating that further improvement in the formulation of SCC-DFTB is still a challenge to meet. To make SCC-DFTB applicable to phosphate reactions in the current (on-site-third-order-only) formulation, a "reaction-specific" parameterization, referred to as SCC-DFTBPR, is developed based on hydrolysis reactions of model phosphate species. Benchmark calculations in both the gas-phase and solution-phase indicate that SCC-DFTBPR gives reliable structural properties and semi-quantitative energetics for phosphate hydrolysis reactions. Since the number of reaction-specific parameters is small, it is likely that SCC-DFTBPR is applicable to a broad set of phosphate species. Indeed, for 56 reaction exothermicities and 47 energy barriers related to RNA catalysis model reactions collected from the QCRNA database, which involve molecules rather different from those used to parameterize SCC-DFTBPR, the corresponding root-mean-square difference between SCC-DFTBPR and high-level DFT results is only 5.3 kcal/mol. We hope that the parameterized SCC-DFTB models will complement NDDO based reaction-specific models (e.g., AM1-d/PhoT) and high-level ab initio QM/MM methods in better understanding the mechanism of phosphate chemistry in condensed phase, particularly biological systems. Read More View Full Article Download PDF |
 | Extension of adaptive tree code and fast multipole methods to high angular momentum particle charge densities Journal of Computational Chemistry (2008) 29, 1895-1904 DOI: 10.1002/jcc.20946 PMC2716046The development and implementation of a tree code (TC) and fast multipole method (FMM) for the efficient, linear-scaling calculation of long-range electrostatic interactions of particle distributions with variable shape and multipole character are described. The target application of these methods are stochastic boundary molecular simulations with polarizable force fields and/or combined quantum mechanical/molecular mechanical potentials. Linear-scaling is accomplished through the adaptive decomposition of the system into a hierarchy of interacting particle sets. Two methods for effecting this decomposition are evaluated: fluc-splitting and box-splitting, for which the latter is demonstrated to be generally more accurate. In addition, a generalized termination criterion is developed that delivers optimal performance at fixed error tolerance that, in the case of quadrupole-represented Drude water, effects a speed-up by a factor of 2–3 relative to a multipole-independent termination criteria. The FMM is shown to be ~2–3 times faster than the TC, independent of the system size and multipole order of the particles. The TC and FMM are tested for a variety of static and polarizable water systems, and for the the 70S ribosome functional complex containing an assembly of transfer and messenger RNAs. Read More View Full Article Download PDF |
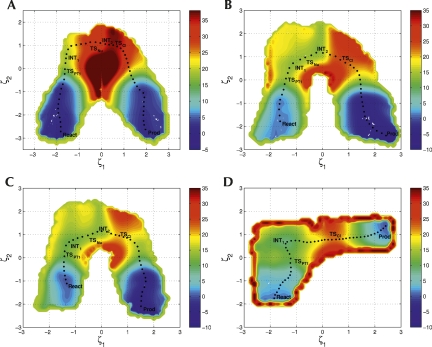 | Electrostatic interactions in the hairpin ribozyme account for the majority of the rate acceleration without chemical participation by nucleobases RNA (2008) 14, 1501-1507 DOI: 10.1261/rna.863108 PMC2491468Molecular dynamics simulations using a combined quantum mechanical/molecular mechanical potential are used to determine the two-dimensional free energy profiles for the mechanism of RNA transphosphorylation in solution and catalyzed by the hairpin ribozyme. A mechanism is explored whereby the reaction proceeds without explicit chemical participation by conserved nucleobases in the active site. The ribozyme lowers the overall free energy barrier by up to 16 kcal/mol, accounting for the majority of the observed rate enhancement. The barrier reduction in this mechanism is achieved mainly by the electrostatic environment provided by the ribozyme without recruitment of active site nucleobases as acid or base catalysts. The results establish a baseline mechanism that invokes only the solvation and specific hydrogen-bonding interactions present in the ribozyme active site and provide a departure point for the exploration of alternate mechanisms where nucleobases play an active chemical role. Read More View Full Article Download PDF |
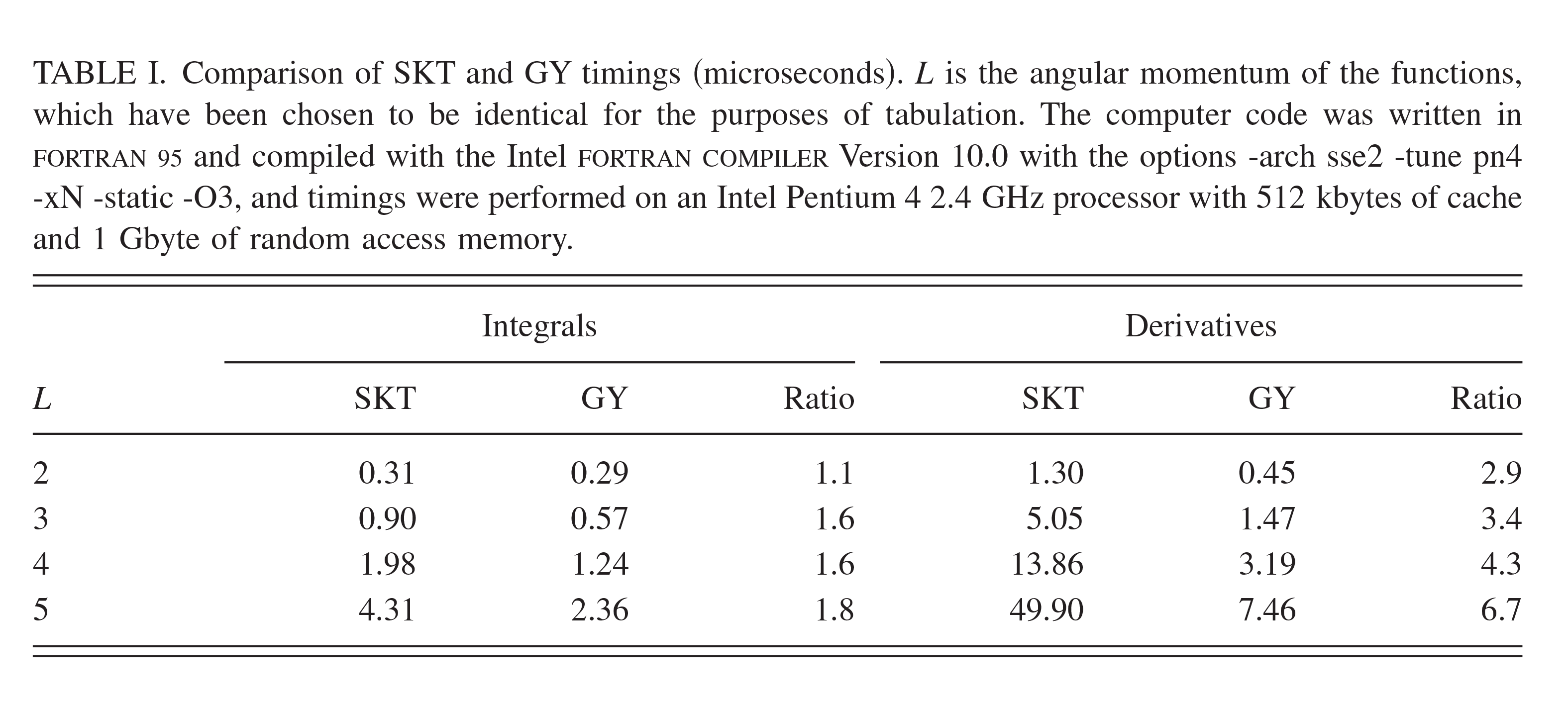 | Spherical tensor gradient operator method for integral rotation: A simple, efficient, and extendable alternative to Slater-Koster tables The Journal of Chemical Physics (2008) 129, 016102 DOI: 10.1063/1.2945897 PMC2671661We present a novel alternative to the use of Slater–Koster tables for the efficient rotation and gradient evaluation of two-center integrals used in tight-binding Hamiltonian models. The method recasts the problem into an exact, yet implicit, basis representation through which the properties of the spherical tensor gradient operator are exploited. These properties provide a factor of 3 to 4 speedup in the evaluation of the integral gradients and afford a compact code structure that easily extends to high angular momentum without loss in efficiency. Thus, the present work is important in improving the performance of tight-binding models in molecular dynamics simulations and has particular use for methods that require the evaluation of two-center integrals that involve high angular momentum basis functions. These advances have a potential impact for the design of new tight-binding models that incorporate polarization or transition metal basis functions and methods based on electron density fitting of molecular fragments. Read More View Full Article Download PDF |
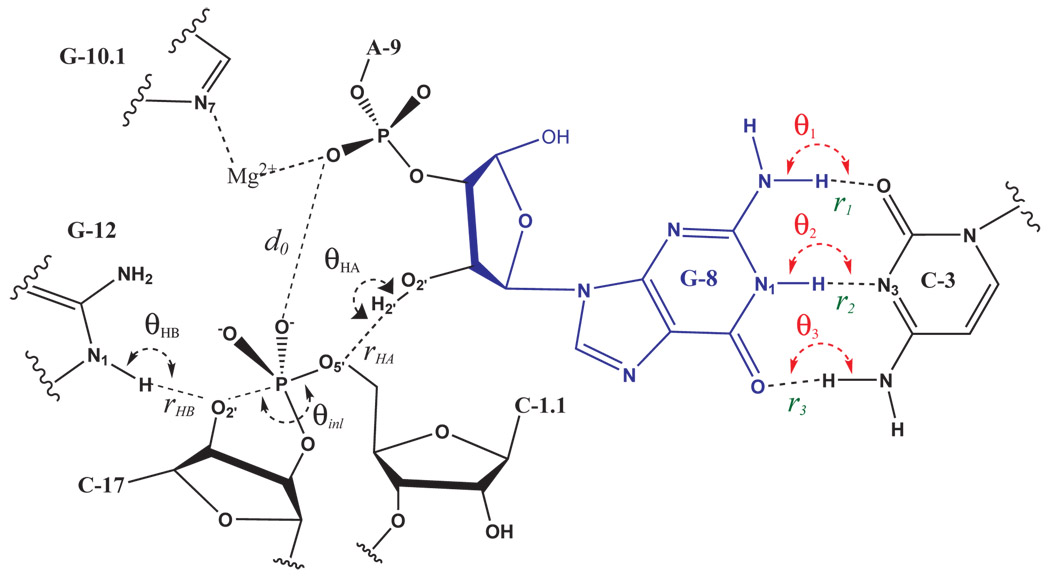 | Origin of Mutational Effects at the C3 and G8 Positions on Hammerhead Ribozyme Catalysis from Molecular Dynamics Simulations Journal of the American Chemical Society (2008) 130, 7168-7169 DOI: 10.1021/ja711242b PMC2733889A series of ten 60 ns molecular dynamics (MD) simulations of the native and mutated full length hammerhead ribozymes in the reactant state and in an activated precursor state (G8:2'OH deprotonated) are reported. Mutant simulations include the C3U, G8A, and G8I single mutants and a C3U/G8A double mutant that exhibits an experimental rescue effect. The results provide critical details into the origin of the observed mutation effects and support a mechanism where the 2'OH of G8 acts as a general acid catalyst that is held in position through Watson-Crick hydrogen bonding between G8 and C3. Read More View Full Article Download PDF |
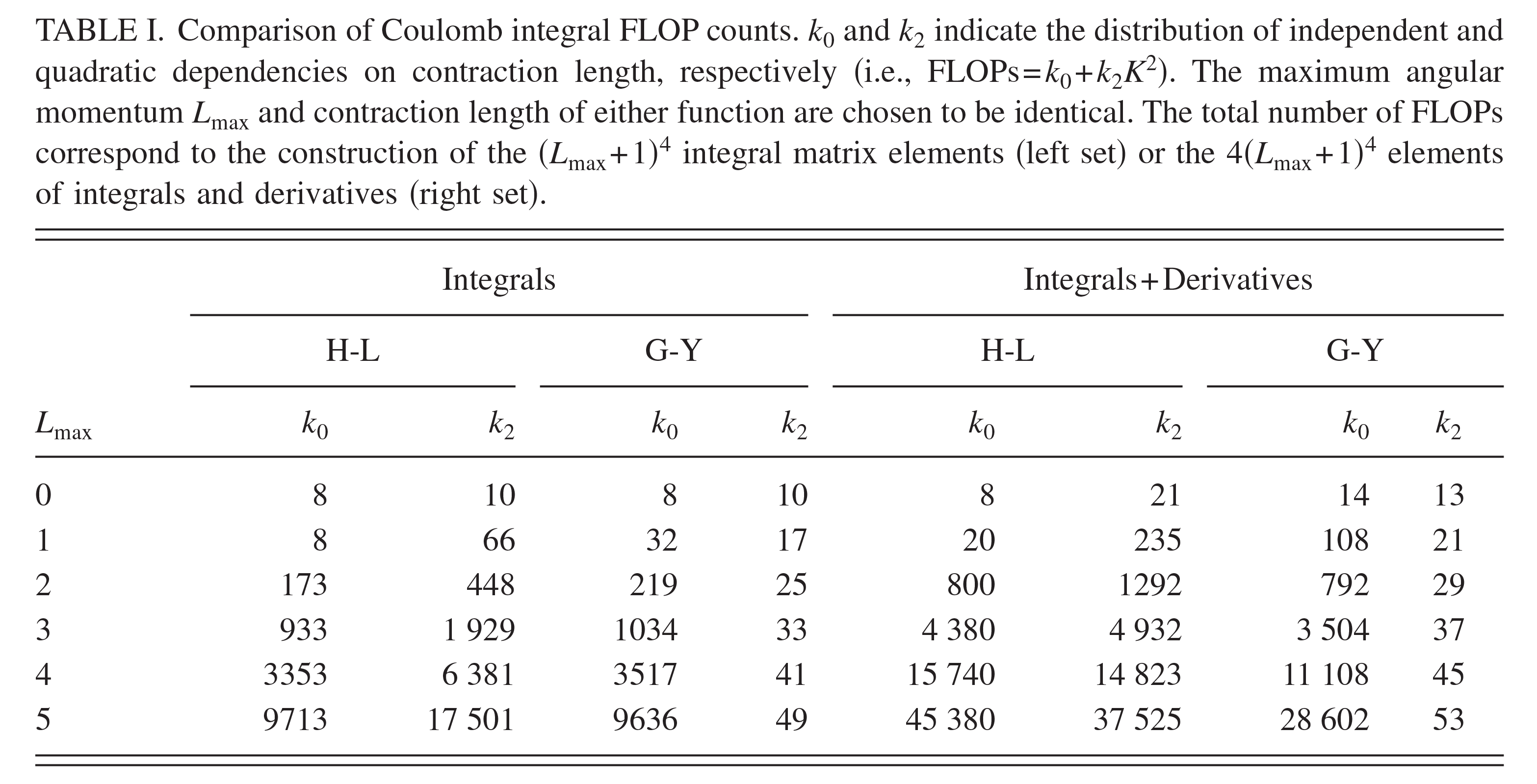 | Contracted auxiliary Gaussian basis integral and derivative evaluation The Journal of Chemical Physics (2008) 128, 064104 DOI: 10.1063/1.2821745 PMC2735875The rapid evaluation of two-center Coulomb and overlap integrals between contracted auxiliary solid harmonic Gaussian functions is examined. Integral expressions are derived from the application of Hobson's theorem and Dunlap's product and differentiation rules of the spherical tensor gradient operator. It is shown that inclusion of the primitive normalization constants greatly simplifies the calculation of contracted functions corresponding to a Gaussian multipole expansion of a diffuse charge density. Derivative expressions are presented and it is shown that chain rules are avoided by expressing the derivatives as a linear combination of auxiliary integrals involving no more than five terms. Calculation of integrals and derivatives requires the contraction of a single vector corresponding to the monopolar result and its scalar derivatives. Implementation of the method is discussed and comparison is made with a Cartesian Gaussian-based method. The current method is superior for the evaluation of both integrals and derivatives using either primitive or contracted functions. Read More View Full Article Download PDF |
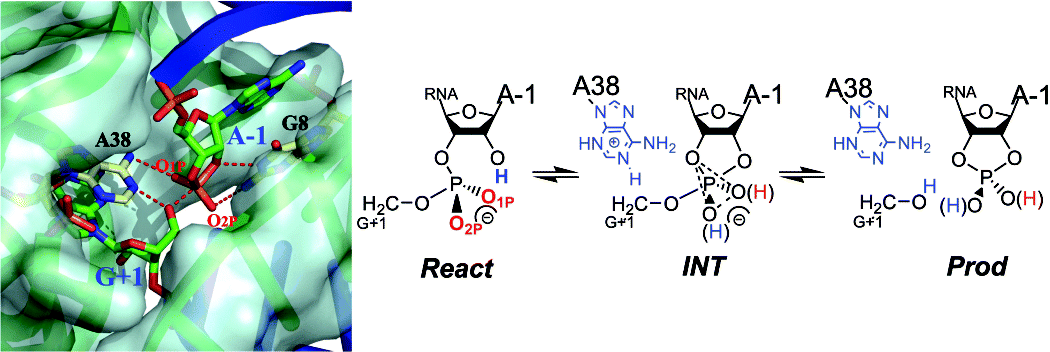 | Quantum mechanical/molecular mechanical simulation study of the mechanism of Hairpin ribozyme catalysis Journal of the American Chemical Society (2008) 130, 4680-4691 DOI: 10.1021/ja0759141 PMC2655239The molecular mechanism of hairpin ribozyme catalysis is studied with molecular dynamics simulations using a combined quantum mechanical and molecular mechanical (QM/MM) potential with a recently developed semiempirical AM1/d-PhoT model for phosphoryl transfer reactions. Simulations are used to derive one- and two-dimensional potentials of mean force to examine specific reaction paths and assess the feasibility of proposed general acid and base mechanisms. Density-functional calculations of truncated active site models provide complementary insight to the simulation results. Key factors utilized by the hairpin ribozyme to enhance the rate of transphosphorylation are presented, and the roles of A38 and G8 as general acid and base catalysts are discussed. The computational results are consistent with available experimental data, provide support for a general acid/base mechanism played by functional groups on the nucleobases, and offer important insight into the ability of RNA to act as a catalyst without explicit participation by divalent metal ions. Read More View Full Article Download PDF |
 | Solvent structure and hammerhead ribozyme catalysis Chemistry & Biology (2008) 15, 332-342 DOI: 10.1016/j.chembiol.2008.03.010 PMC2610685Although the hammerhead ribozyme is regarded as a prototype for understanding RNA catalysis, the mechanistic roles of associated metal ions and water molecules in the cleavage reaction remain controversial. We have investigated the catalytic potential of observed divalent metal ions and water molecules bound to a 2 A structure of the full-length hammerhead ribozyme by using X-ray crystallography in combination with molecular dynamics simulations. A single Mn2+ is observed to bind directly to the A9 phosphate in the active site, accompanying a hydrogen-bond network involving a well-ordered water molecule spanning N1 of G12 (the general base) and 2'-O of G8 (previously implicated in general acid catalysis) that we propose, based on molecular dynamics calculations, facilitates proton transfer in the cleavage reaction. Phosphate-bridging metal interactions and other mechanistic hypotheses are also tested with this approach. Read More View Full Article Download PDF |
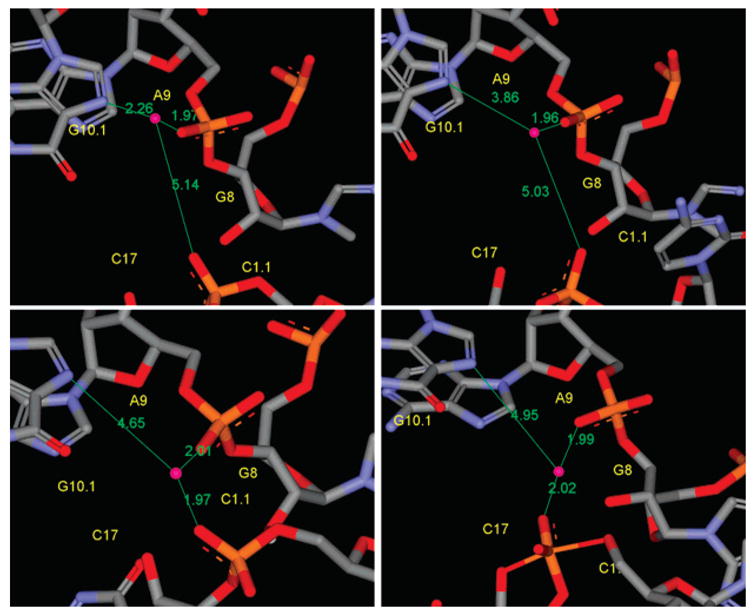 | Role of Mg2+ in hammerhead ribozyme catalysis from molecular simulation Journal of the American Chemical Society (2008) 130, 3053-3064 DOI: 10.1021/ja076529e PMC2535817Molecular dynamics simulations have been performed to investigate the role of Mg2+ in the full-length hammerhead ribozyme cleavage reaction. In particular, the aim of this work is to characterize the binding mode and conformational events that give rise to catalytically active conformations and stabilization of the transition state. Toward this end, a series of eight 12 ns molecular dynamics simulations have been performed with different divalent metal binding occupations for the reactant, early and late transition state using recently developed force field parameters for metal ions and reactive intermediates in RNA catalysis. In addition, hybrid QM/MM calculations of the early and late transition state were performed to study the proton-transfer step in general acid catalysis that is facilitated by the catalytic Mg2+ ion. The simulations suggest that Mg2+ is profoundly involved in the hammerhead ribozyme mechanism both at structural and catalytic levels. Binding of Mg2+ in the active site plays a key structural role in the stabilization of stem I and II and to facilitate formation of near attack conformations and interactions between the nucleophile and G12, the implicated general base catalyst. In the transition state, Mg2+ binds in a bridging position where it stabilizes the accumulated charge of the leaving group while interacting with the 2'OH of G8, the implicated general acid catalyst. The QM/MM simulations provide support that, in the late transition state, the 2'OH of G8 can transfer a proton to the leaving group while directly coordinating the bridging Mg2+ ion. The present study provides evidence for the role of Mg2+ in hammerhead ribozyme catalysis. The proposed simulation model reconciles the interpretation of available experimental structural and biochemical data, and provides a starting point for more detailed investigation of the chemical reaction path with combined QM/MM methods. Read More View Full Article Download PDF |
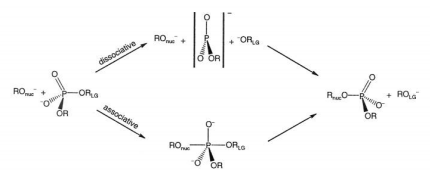 | New QM/MM Models for Multi-scale Simulation of Phosphoryl Transfer Reactions in Solution Multiscale Simulation Methods for Nanomaterials(2007) 201-218 Chapter: Multiscale Simulation Methods for Nanomaterials DOI: 10.1002/9780470191675.ch12 ISBN: 9780470191675This book stems from the American Chemical Society symposium, Large Scale Molecular Dynamics, Nanoscale, and Mesoscale Modeling and Simulation: Bridging the Gap, that delved into the latest methodologies and applications for largescale, multiscale, and mesoscale modeling and simulation. It presents real-world applications of simulated and synthesized materials, including organic-, inorganic-, bio-, and nanomaterials, and helps readers determine the best method for their simulation. It gets novices up to speed quickly and helps experienced practitioners discover novel approaches and alternatives. Read More View Full Article Download PDF |
 | Charge-dependent model for many-body polarization, exchange and dispersion interactions in hybrid QM/MM calculations The Journal of Chemical Physics (2007) 127, 194101 DOI: 10.1063/1.2778428 18035873This work explores a new charge-dependent energy model consisting of van der Waals and polarization interactions between the quantum mechanical (QM) and molecular mechanical (MM) regions in a combined QMMM calculation. van der Waals interactions are commonly treated using empirical Lennard-Jones potentials, whose parameters are often chosen based on the QM atom type (e.g., based on hybridization or specific covalent bonding environment). This strategy for determination of QMMM nonbonding interactions becomes tedious to parametrize and lacks robust transferability. Problems occur in the study of chemical reactions where the "atom type" is a complex function of the reaction coordinate. This is particularly problematic for reactions, where atoms or localized functional groups undergo changes in charge state and hybridization. In the present work we propose a new model for nonelectrostatic nonbonded interactions in QMMM calculations that overcomes many of these problems. The model is based on a scaled overlap model for repulsive exchange and attractive dispersion interactions that is a function of atomic charge. The model is chemically significant since it properly correlates atomic size, softness, polarizability, and dispersion terms with minimal one-body parameters that are functions of the atomic charge. Tests of the model are examined for rare-gas interactions with neutral and charged atoms in order to demonstrate improved transferability. The present work provides a new framework for modeling QMMM interactions with improved accuracy and transferability. Read More View Full Article Download PDF |
 | Extension of the self-consistent-charge density-functional tight-binding method: third-order expansion of the density functional theory total energy and introduction of a modified effective coulomb interaction The Journal of Physical Chemistry A (2007) 111, 10861-10873 DOI: 10.1021/jp074167r 17914769The standard self-consistent-charge density-functional-tight-binding (SCC-DFTB) method (Phys. Rev. B 1998, 58, 7260) is derived by a second-order expansion of the density functional theory total energy expression, followed by an approximation of the charge density fluctuations by charge monopoles and an effective damped Coulomb interaction between the atomic net charges. The central assumptions behind this effective charge-charge interaction are the inverse relation of atomic size and chemical hardness and the use of a fixed chemical hardness parameter independent of the atomic charge state. While these approximations seem to be unproblematic for many covalently bound systems, they are quantitatively insufficient for hydrogen-bonding interactions and (anionic) molecules with localized net charges. Here, we present an extension of the SCC-DFTB method to incorporate third-order terms in the charge density fluctuations, leading to chemical hardness parameters that are dependent on the atomic charge state and a modification of the Coulomb scaling to improve the electrostatic treatment within the second-order terms. These modifications lead to a significant improvement in the description of hydrogen-bonding interactions and proton affinities of biologically relevant molecules. Read More View Full Article Download PDF |
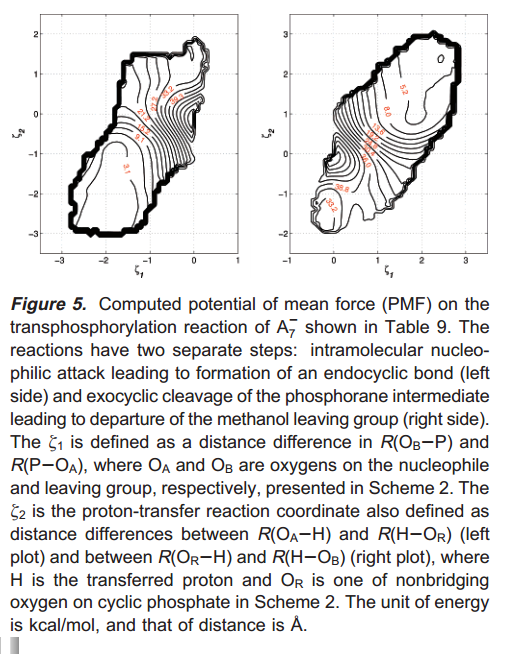 | Specific reaction parametrization of the AM1/d Hamiltonian for phosphoryl transfer reactions: H, O, and P atoms Journal of Chemical Theory and Computation (2007) 3, 486-504 DOI: 10.1021/ct6002466 26637030A semiempirical AM1/d Hamiltonian is developed to model phosphoryl transfer reactions catalyzed by enzymes and ribozymes for use in linear-scaling calculations and combined quantum mechanical/molecular mechanical simulations. The model, designated AM1/d-PhoT, is parametrized for H, O, and P atoms to reproduce high-level density-functional results from a recently constructed database of quantum calculations for RNA catalysis, including geometries and relative energies of minima, transition states and reactive intermediates, dipole moments, proton affinities, and other relevant properties. The model is tested in the gas phase and in solution using a QM/MM potential. The results indicate that the method provides significantly higher accuracy than MNDO/d, AM1, and PM3 methods and, for the transphosphorylation reactions, is in close agreement with the density-functional calculations at the B3LYP/6-311++G(3df,2p) level with a reduction in computational cost of 3-4 orders of magnitude. The model is expected to have considerable impact on the application of semiempirical QM/MM methods to transphosphorylation reactions in solution, enzymes, and ribozymes and to ultimately facilitate the design of improved next-generation multiscale quantum models. Read More View Full Article Download PDF |
 | Insight into the role of Mg2+in hammerhead ribozyme catalysis from x-ray crystallography and molecular dynamics simulation Journal of Chemical Theory and Computation (2007) 3, 325-327 DOI: 10.1021/ct6003142 PMC2600717Results of a series of 12 ns molecular dynamics (MD) simulations of the reactant state (with and without a Mg2+ ion), early and late transition state mimics are presented based on a recently reported crystal structure of a full-length hammerhead RNA. The simulation results support a catalytically active conformation with a Mg2+ ion bridging the A9 and scissile phosphates. In the reactant state, the Mg2+ spends significant time closely associated with the 2'OH of G8, but remains fairly distant from the leaving group O(5') position. In the early TS mimic simulation, where the nucleophilic O(2') and leaving group O(5') are equidistant from the phosphorus, the Mg2+ ion remains tightly coordinated to the 2'OH of G8, but is positioned closer to the O(5') leaving group, stabilizing the accumulating charge. In the late TS mimic simulation, the coordination around the bridging Mg2+ ion undergoes a transition whereby the coordination with the 2'OH of G8 is replace by the leaving group O(5') that has developed significant charge. At the same time, the 2'OH of G8 forms a hydrogen bond with the leaving group O(5') and is positioned to act as a general acid catalyst. This work represents the first reported simulations of the full-length hammerhead structure and TS mimics, and provides direct evidence for the possible role of a bridging Mg2+ ion in catalysis that is consistent with both crystallographic and biochemical data. Read More View Full Article Download PDF |
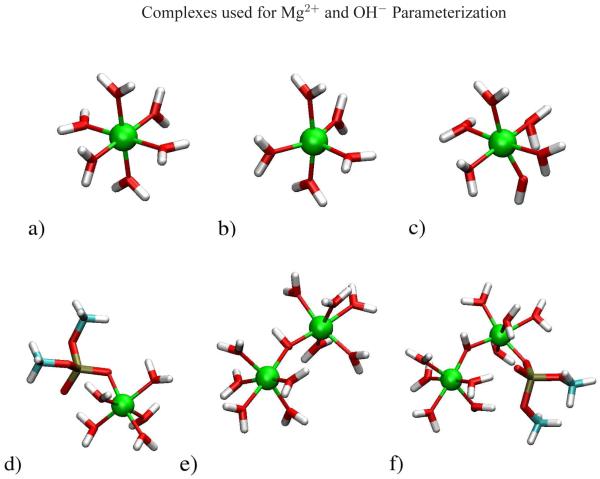 | CHARMM force field parameters for simulation of reactive intermediates in native and thio-substituted ribozymes Journal of Computational Chemistry (2007) 28, 495-507 DOI: 10.1002/jcc.20474 PMC2869295Force field parameters specifically optimized for residues important in the study of RNA catalysis are derived from density-functional calculations, in a fashion consistent with the CHARMM27 all-atom empirical force field. Parameters are presented for residues that model reactive RNA intermediates and transition state analogs, thio-substituted phosphates and phosphoranes, and bound Mg2+ and di-metal bridge complexes. Target data was generated via density-functional calculations at the B3LYP/6–311++G(3df,2p)// B3LYP/6–31++G(d,p) level. Partial atomic charges were initially derived from CHelpG electrostatic potential fitting and subsequently adjusted to be consistent with the CHARMM27 charges. Lennard-Jones parameters were determined to reproduce interaction energies with water molecules. Bond, angle, and torsion parameters were derived from the density-functional calculations and renormalized to maintain compatibility with the existing CHARMM27 parameters for standard residues. The extension of the CHARMM27 force field parameters for the nonstandard biological residues presented here will have considerable use in simulations of ribozymes, including the study of freeze-trapped catalytic intermediates, metal ion binding and occupation, and thio effects. Read More View Full Article Download PDF |
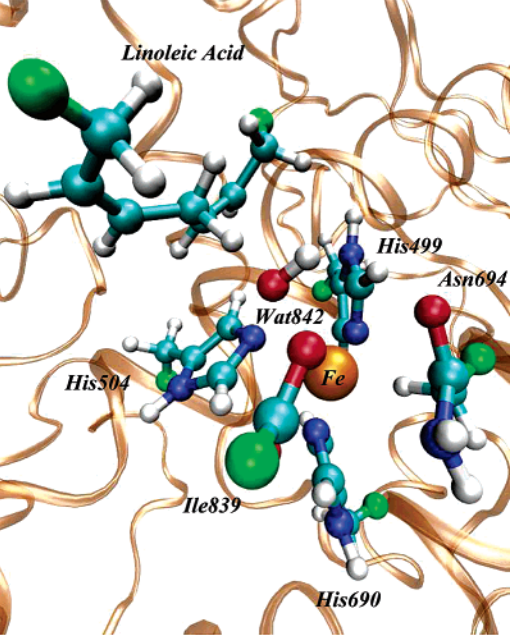 | Enzyme dynamics and tunneling enhanced by compression in the hydrogen abstraction catalyzed by soybean lipoxygenase-1 The Journal of Physical Chemistry B (2006) 110, 24708-24719 DOI: 10.1021/jp066263i 17134234A fully microscopical simulation of the rate-limiting hydrogen abstraction catalyzed by soybean lipoxygenase-1 (SLO-1) has been carried out. This enzyme exhibits the largest, and weakly temperature dependent, experimental H/D kinetic isotope effect (KIE) reported for a biological system. The theoretical model used here includes the complete enzyme with a solvation shell of water molecules, the Fe(III)-OH- cofactor, and the linoleic acid substrate. We have used a hybrid QM(PM3/d-SRP)/MM method to describe the potential energy surface of the whole system, and the ensemble-averaged variational transition-state theory with multidimensional tunneling (EA-VTST/MT) to calculate the rate constant and the primary KIE. The computational results show that the compression of the wild-type active site enzyme results in the huge contribution of tunneling (99%) to the rate of the hydrogen abstraction. Importantly, the active site becomes more flexible in the Ile553Ala mutant reactant complex simulation (for which a markedly temperature dependent KIE has been experimentally determined), thus justifying the proposed key role of the gating promoting mode in the reaction catalyzed by SLO-1. Finally, the results indicate that the calculated KIE for the wild-type enzyme has an important dependence on the barrier width. Read More View Full Article Download PDF |
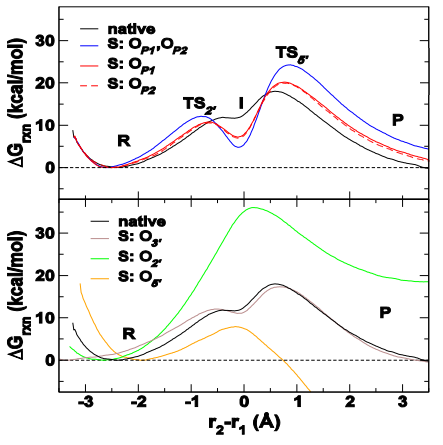 | Simulations of phosphoryl transfer reactions using multi-scale quantum models Modelling Molecular Structure and Reactivity in Biological Systems(2006) 181-192 DOI: 10.1039/9781847555373 Computational and theoretical tools for understanding biological processes at the molecular level is an exciting and innovative area of science. Using these methods to study the structure, dynamics and reactivity of biomacromolecules in solution, computational chemistry is becoming an essential tool, complementing the more traditional methods for structure and reactivity determination. Modelling Molecular Structure and Reactivity in Biological Systems covers three main areas in computational chemistry; structure (conformational and electronic), reactivity and design. Initial sections focus on the link between computational and spectroscopic methods in the investigation of electronic structure. The use of Free Energy calculations for the elucidation of reaction mechanisms in enzymatic systems is also discussed. Subsequent sections focus on drug design and the use of database methods to determine ADME (absorption, distribution, metabolism, excretion) properties. This book provides a complete reference on state of the art computational chemistry practised on biological systems. It is ideal for researchers in the field of computational chemistry interested in its application to biological systems. Read More View Full Article Download PDF |
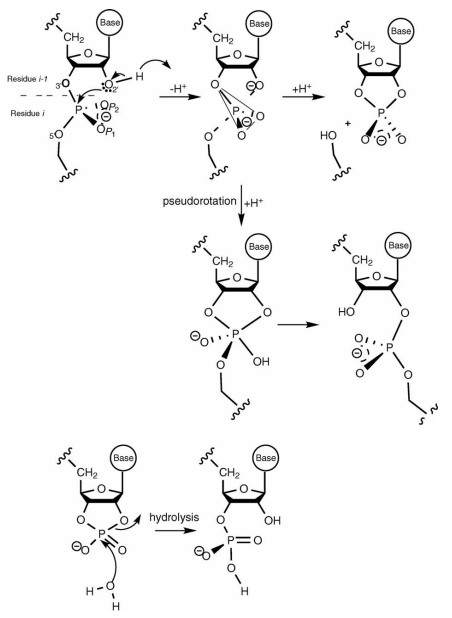 | QCRNA 1.0: A database of quantum calculations for RNA catalysis Journal of Molecular Graphics and Modelling (2006) 25, 423-433 DOI: 10.1016/j.jmgm.2006.02.011 16580853This work outlines a new on-line database of quantum calculations for RNA catalysis (QCRNA) available via the worldwide web at http://theory.rutgers.edu/QCRNA/. The database contains high-level density functional calculations for a large range of molecules, complexes and chemical mechanisms important to phosphoryl transfer reactions and RNA catalysis. Calculations are performed using a strict, consistent protocol such that a wealth of cross-comparisons can be made to elucidate meaningful trends in biological phosphate reactivity. Currently, around 2000 molecules have been collected in varying charge states in the gas phase and in solution. Solvation was treated with both the PCM and COSMO continuum solvation models. The data can be used to study important trends in reactivity of biological phosphates, or used as benchmark data for the design of new semiempirical quantum models for hybrid quantum mechanical/molecular mechanical simulations. Read More View Full Article Download PDF |
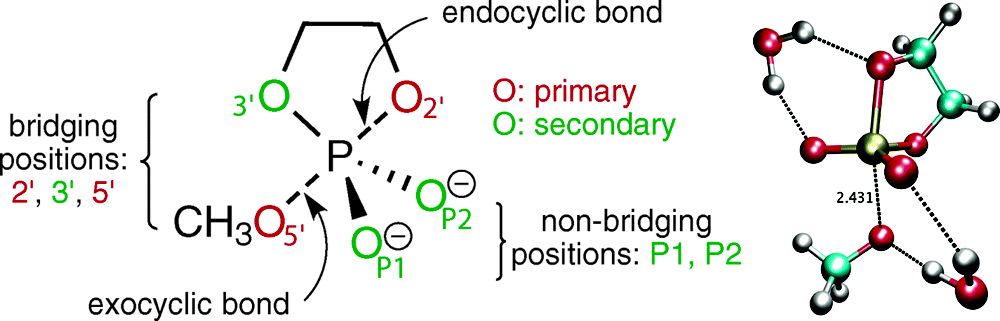 | Transesterification thio effects of phosphate diesters: free energy barriers and kinetic and equilibrium isotope effects from density-functional theory Biochemistry (2006) 45, 10043-10053 DOI: 10.1021/bi060869f 16906762Primary and secondary kinetic and equilibrium isotope effects are calculated with density-functional methods for the in-line dianionic methanolysis of the native (unsubstituted) and thio-substituted cyclic phosphates. These reactions represent reverse reaction models for RNA transesterification under alkaline conditions. The effect of solvent is treated with explicit (single and double) water molecules and self-consistently with an implicit (continuum) solvation model. Singly substituted reactions at the nonbridging OP1 position and bridging O2‘, O3‘, and O5‘ positions and a doubly substituted reaction at the OP1 and OP2 positions were considered. Aqueous free energy barriers are calculated, and the structures and bond orders of the rate-controlling transition states are characterized. The results are consistent with available experimental data and provide useful information for the interpretation of measured isotope and thio effects used to probe mechanism in phosphoryl transfer reactions catalyzed by enzymes and ribozymes. Read More View Full Article Download PDF |
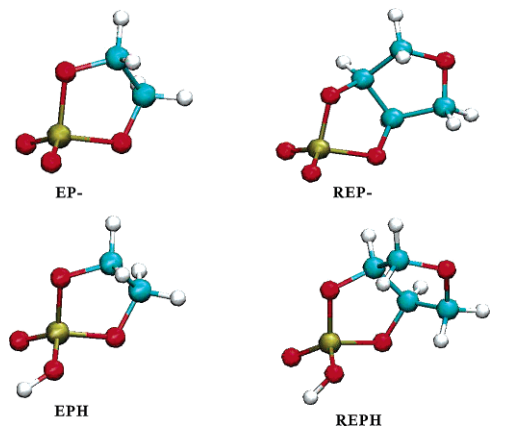 | Nucleophilic attack on phosphate diesters: a density functional study of in-line reactivity in dianionic, monoanionic, and neutral systems The Journal of Physical Chemistry B (2006) 110, 11525-11539 DOI: 10.1021/jp0603942 16771429A density functional study of the hydrolysis reaction of phosphodiesters with a series of attacking nucleophiles in the gas phase and in solution is presented. The nucleophiles HOH, HO-, CH3OH, and CH3O- were studied in reactions with ethylene phosphate, 2‘3‘-ribose cyclic phosphate and in their neutral (protonated) and monoanionic forms. Stationary-point geometries for the reactions were determined at the density functional B3LYP/6-31++G(d,p) level followed by energy refinement at the B3LYP/6-311++G(3df,2p) level. Solvation effects were estimated by using a dielectric approximation with the polarizable continuum model (PCM) at the gas-phase optimized geometries. This series of reactions characterizes factors that influence the intrinsic reactivity of the model phosphate compounds, including the effect of nucleophile, protonation state, cyclic structure, and solvent. The present study of the in-line mechanism for phosphodiester hydrolysis, a reaction of considerable biological importance, has implications for enzymatic mechanisms. The analysis generally supports the associative mechanism for phosphate ester hydrolysis. The results highlight the importance for the reaction barrier of charge neutralization resulting from the protonation of the nonbridging phosphoryl oxygens and the role of internal hydrogen transfer in the gas-phase mechanism. It also shows that solvent stabilization has a profound influence on the relative barrier heights for the dianionic, monoanionic, and neutral reactions. The calculations provide a comprehensive data set for the in-line hydrolysis mechanisms that can be used for the development of improved semiempirical quantum models for phosphate hydrolysis reactions. Read More View Full Article Download PDF |
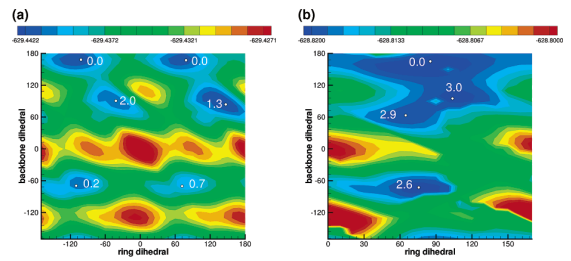 | Normal modes of redox-active tyrosine: conformation dependence and comparison to experiment The Journal of Physical Chemistry B (2006) 110, 10970-10981 DOI: 10.1021/jp061503f 16771350Redox-active tyrosine residues play important roles in long-distance electron reactions in enzymes such as prostaglandin H synthase, ribonucleotide reductase, and photosystem II (PSII). Spectroscopic characterization of tyrosyl radicals in these systems provides a powerful experimental probe into the role of the enzyme in mediation of long-range electron transfer processes. Interpretation of such data, however, relies critically on first establishing a spectroscopic fingerprint of isotopically labeled tyrosinate and tyrosyl radicals in nonenzymatic environments. In this report, FT-IR results obtained from tyrosinate, tyrosyl radical (produced by ultraviolet photolysis of polycrystalline tyrosinate), and their isotopologues at 77 K are presented. Assignment of peaks and isotope shifts is aided by density-functional B3LYP/6-311++G(3df,2p)//B3LYP/6-31++G(d,p) calculations of tyrosine and tyrosyl radical in several different charge and protonation states. In addition, characterization of the potential energy surfaces of tyrosinate and tyrosyl radical as a function of the backbone and ring torsion angles provides detailed insight into the sensitivity of the vibrational frequencies to conformational changes. These results provide a detailed spectroscopic interpretation, which will elucidate the structures of redox-active tyrosine residues in complex protein environments. Specific application of these data is made to enzymatic systems. Read More View Full Article Download PDF |
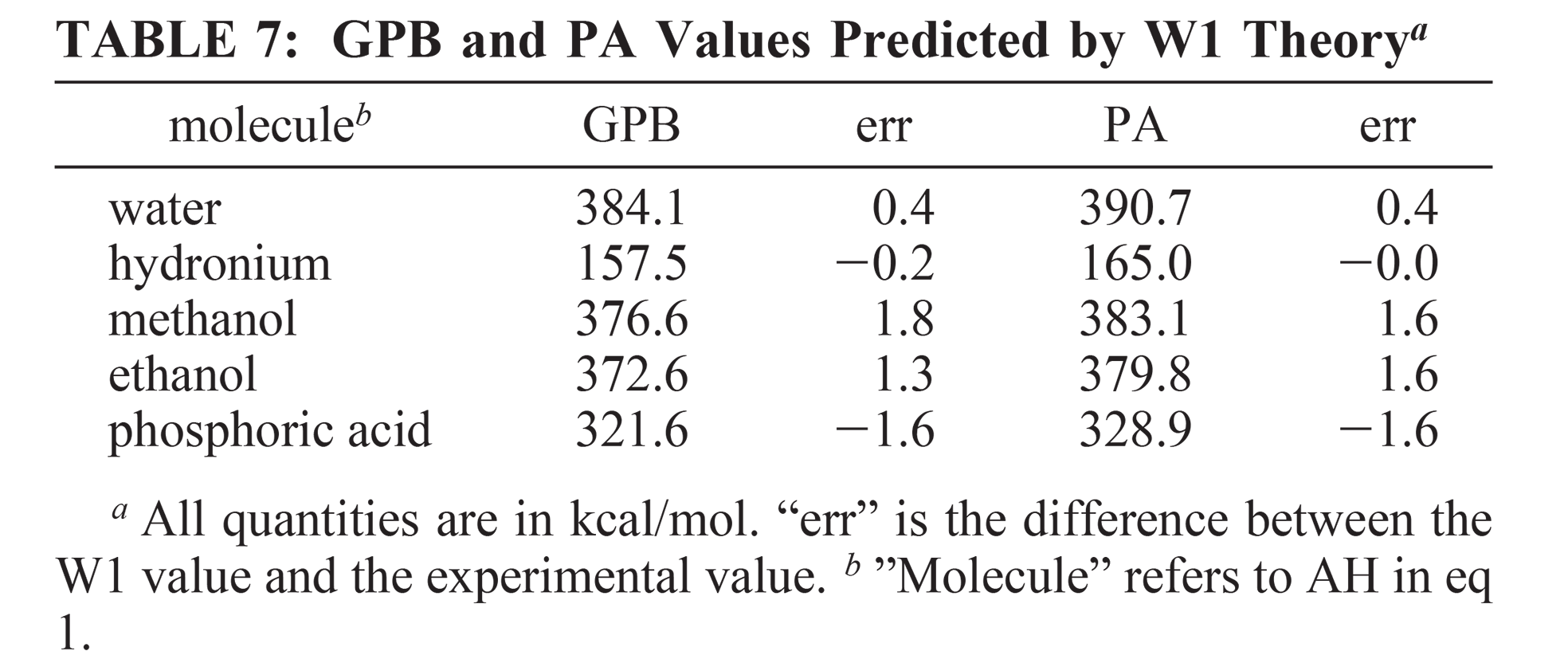 | Multi-level and density functional electronic structure calculations of proton affinities and gas-phase basicities involved in biological phosphoryl transfer The Journal of Physical Chemistry A (2006) 110, 791-797 DOI: 10.1021/jp054360q 16405355Five multilevel model chemistries (CBS-QB3, G3B3, G3MP2B3, MCG3/3, and MC-QCISD/3) and seven hybrid density functional methods (PBE0, B1B95, B3LYP, MPW1KCIS, PBE1KCIS, and MPW1B95) have been applied to the calculation of gas-phase basicity and proton affinity values for a series of 17 molecules relevant to the study of biological phosphoryl transfer. In addition, W1 calculations were performed on a subset of molecules. The accuracy of the methods was assessed and the nature of systematic errors was explored, leading to the introduction of a set of effective bond enthalpy and entropy correction terms. The multicoefficient correlation methods (MCG3/3 and MC-QCISD), with inclusion of specific zero-point scale factors, slightly outperform the other multilevel methods tested (CBS-QB3, G3B3, and G3MP2B3), with significantly less computational cost, and in the case of MC-QCISD, slightly less severe scaling. Four density functional methods, PBE1KCIS, MPW1B95, PBE0, and B1B95 perform nearly as well as the multilevel methods. These results provide an important set of benchmarks relevant to biological phosphoryl transfer reactions. Read More View Full Article Download PDF |
 | A charge-scaling implementation of the variational electrostatic projection method Journal of Computational Chemistry (2006) 27, 103-115 DOI: 10.1002/jcc.20318 16273506Two new charge-scaling methods for efficient modeling of the solvated macromolecular environment in hybrid QM/MM calculations of biological reactions are presented. The methods are extensions of the variational electrostatic projection (VEP) method, and allows a subset of atomic charges in the external environment to be adjusted to mimic, in the active dynamical region, the electrostatic potential and field due to the large surrounding macromolecule and solvent. The method has the advantages that it offers improved accuracy, does not require the use of a three-dimensional grid or auxiliary set of fitting points, and requires only minor molecular simulation code modifications. The VEP-cs and VEP-RVM+cs methods are able to attain very high accuracy (relative force errors of 10-7 or better with appropriate choice of control parameters), and take advantage of a recently introduced set of high-order discretization schemes and Gaussian exponents for boundary element solvation and VEP methods. The methods developed here serve as potentially powerful tools in the arsenal of computational techniques used in multiscale computational modeling problems. Read More View Full Article Download PDF |
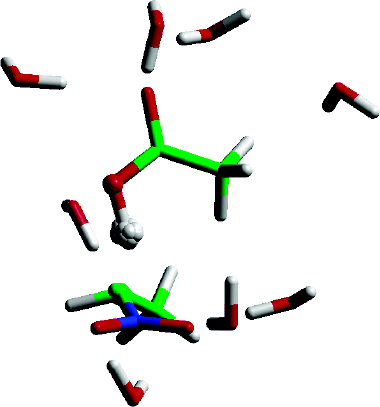 | Solvent polarization and kinetic isotope effects in nitroethane deprotonation and implications to the nitroalkane oxidase reaction Journal of the American Chemical Society (2005) 127, 16374-16375 DOI: 10.1021/ja055881u 16305206We have carried out a mixed molecular dynamics and centroid path integral simulation using a combined quantum mechanical and molecular mechanical (QM/MM) potential to study the anomalous Brønsted relationship between rates and equilibria for deprotonation of nitroalkanes in water, which is known as the nitroalkane anomaly. The deprotonation process is catalyzed by nitroalkane oxidase. Our results show that the difference in solvent polarization effects for the TS and products is a major factor for the differential solvent effects on rate and equilibrium of nitroalkane deprotonation. This is due to poor charge delocalization as a result of slow rehybridization compared to bond breaking. Although solvent effects do not affect significantly the computed kinetic isotope effects in comparison with the gas-phase value, there is slight solvent-induced increase in tunneling. The present results suggest that an effective means by which the transition state can be stabilized in the enzyme nitroalkane oxidase is to facilitate the Ca rehybridization. Read More View Full Article Download PDF |
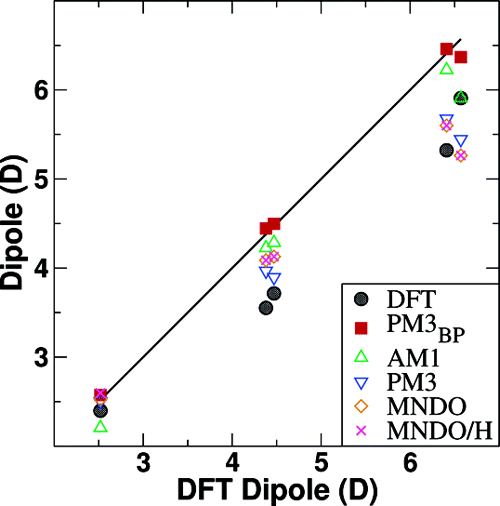 | A semiempirical quantum model for hydrogen bonded nucleic acid base pairs Journal of Chemical Theory and Computation (2005) 1, 1275-1285 DOI: 10.1021/ct050102l 26631671An exploratory semiempirical Hamiltonian (PM3BP) is developed to model hydrogen bonding in nucleic acid base pairs. The PM3BP Hamiltonian is a novel reparametrization of the PM3 Hamiltonian designed to reproduce experimental base pair dimer enthalpies and high-level density-functional results. The parametrization utilized a suite of integrated nonlinear optimization algorithms interfaced with a d-orbital semiempirical program. Results are compared with experimental values and with benchmark density-functional (mPWPW91/MIDI!) calculations for hydrogen-bonded nucleic acid dimers and trimers. The PM3BP Hamiltonian is demonstrated to outperform the AM1, PM3, MNDO, and MNDO/H Hamiltonians for dimer and trimer structures and interaction enthalpies and is shown to reproduce experimental dimer interaction enthalpies that rival density-functional results for an over 3 orders of magnitude reduction in computational cost. The tradeoff between a high accuracy gain for hydrogen bonding at the expense of sacrificing some generality is discussed. These results provide insight into the limits of conventional semiempirical forms for accurate modeling of biological interactions. Read More View Full Article Download PDF |
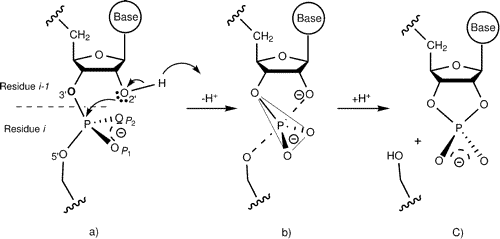 | Density-functional study of the in-line mechanism of methanolysis of cyclic phosphate and thiophosphate esters in solution: insight into thio effects in RNA transesterification The Journal of Physical Chemistry B (2005) 109, 19987-20003 DOI: 10.1021/jp053146z 16853584Density functional calculations of thio effects on the in-line mechanism of methanolysis of ethylene phosphate (a reverse reaction model for RNA phosphate transesterification) are presented. A total of 12 reaction mechanisms are examined using the B3LYP functional with large basis sets, and the effects of solvation were treated using the PCM, CPCM, and SM5 solvation models. Single thio substitutions at all of the distinct phosphoryl oxygen positions (2‘, 3‘, 5‘, pro-R) and a double thio substitution at the nonbridging (pro-R/pro-S) positions were considered. Profiles for each reaction were calculated in the dianionic and monoanionic/monoprotic states, corresponding to reaction models under alkaline and nonalkaline conditions, respectively. These models provide insight into the mechanisms of RNA transesterification thio effects and serve as a set of high-level quantum data that can be used in the design of new semiempirical quantum models for hybrid quantum mechanical/molecular mechanical simulations and linear-scaling electronic structure calculations. Read More View Full Article Download PDF |
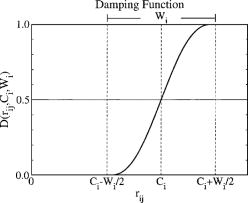 | Improvement of semiempirical response properties with charge-dependent response density The Journal of Chemical Physics (2005) 123, 164108 DOI: 10.1063/1.2080007 16268682The present work outlines a new method for treatment of charge-dependentpolarizability in semiempirical quantum models for use in combined quantum-mechanical/molecular mechanical simulations of biological reactions. The method addresses a major shortcoming in the performance of conventional semiempirical models for these simulations that is tied to the use of a localized minimal atomic-orbital basis set. The present approach has the advantages that it uses a density basis that retains a set of linear-response equations, does not increase the atomic-orbital basis, and avoids the problem of artificial charge transfer and scaling of the polarizability seen in related models that allow atomic charges to fluctuate. The model introduces four new atom-based parameters and has been tested with the modified neglect of differential overlap d-orbital Hamiltonian against 1132molecules and ions and shown to decrease the dipole moment and polarizability errors by factors of 2 and 10, respectively, with respect to density-functional results. The method performs impressively for a variety of charge states (from 2+ to 2-), and offers a potentially powerful extension in the design of next generation semiempirical quantum models for accurate simulations of highly charged biological reactions. Read More View Full Article Download PDF |
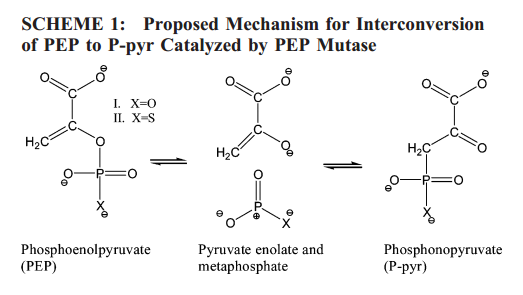 | Theoretical studies of dissociative phosphoryl transfer in interconversion of phosphoenolpyruvate to phosphonopyruvate: solvent effects, thio effects, and implications for enzymatic reactions The Journal of Physical Chemistry B (2005) 109, 13827-13834 DOI: 10.1021/jp051042i 16852731The conversion of phosphoenolpyruvate (PEP) to phosphonopyruvate (P-pyr) is catalyzed by PEP mutase via a dissociative mechanism. In this work, we investigate the uncatalyzed reaction using ab initio methods, density functional theory, and the semiempirical MNDO/d method. Comparisons of geometries and relative energies of stationary points (minima and transition states) with density functional results indicate that the semiempirical method is reasonably accurate. Solvent effects are examined using implicit solvent models, including the recently extended smooth conductor-like screening model. Due to the large negative charge carried by the system, solvation is found to drastically alter the location and energy of stationary points along the dissociative reaction pathways. The influence of substituting a nonbridging phosphoryl oxygen by sulfur (thio effects) was also investigated. Implications of these results for the enzymatic reaction are discussed. Read More View Full Article Download PDF |
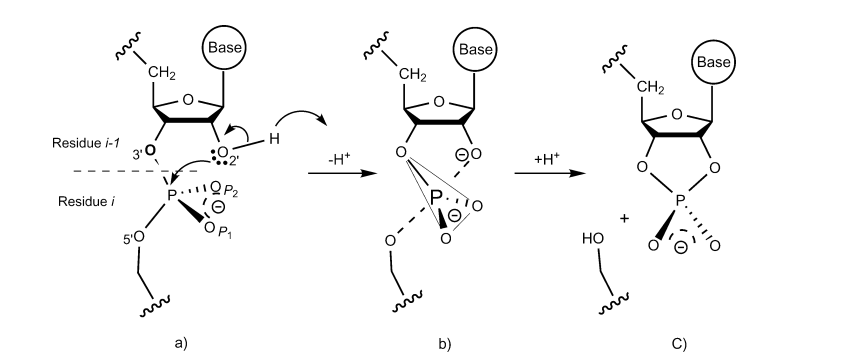 | Benchmark calculations of proton affinities and gas-phase basicities of molecules important in the study of biological phosphoryl transfer Physical Chemistry Chemical Physics (2005) 7, 3070-3079 DOI: 10.1039/B504941E 16186912Benchmark calculations of proton affinities and gas-phase basicities of molecules most relevant to biological phosphoryl transfer reactions are presented and compared with available experimental results. The accuracy of proton affinity and gas-phase basicity results obtained from several multi-level model chemistries (CBS-QB3, G3B3, and G3MP2B3) and density-functional quantum models (PBE0, B1B95, and B3LYP) are assessed and compared. From these data, a set of empirical bond enthalpy, entropy, and free energy corrections are introduced that considerably improve the accuracy and predictive capability of the methods. These corrections are applied to the prediction of proton affinity and gas-phase basicity values of important biological phosphates and phosphoranes for which experimental data does not currently exist. Comparison is made with results from semiempirical quantum models that are commonly employed in hybrid quantum mechanical/molecular mechanical simulations. Data suggest that the design of improved semiempirical quantum models with increased accuracy for relative proton affinity values is necessary to obtain quantitative accuracy for phosphoryl transfer reactions in solution, enzymes, and ribozymes. Read More View Full Article Download PDF |
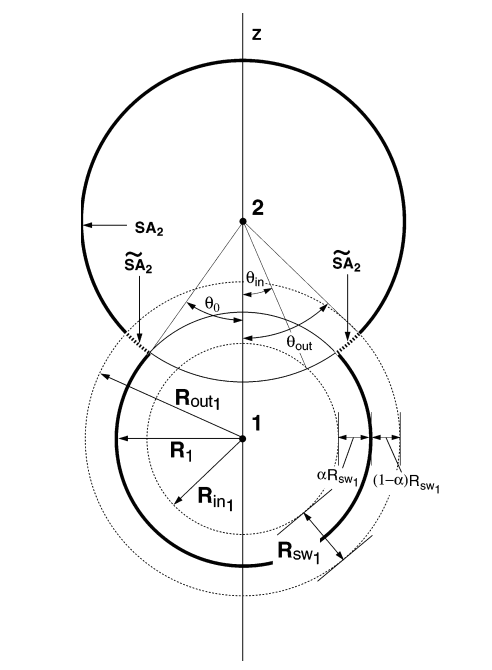 | Smooth solvation method for d-orbital semiempirical calculations of biological reactions. 1. Implementation The Journal of Physical Chemistry B (2005) 109, 9799-9809 DOI: 10.1021/jp044062d 16852180The present paper describes the extension of a recently developed smooth conductor-like screening model for solvation to a d-orbital semiempirical framework (MNDO/d-SCOSMO) with analytic gradients that can be used for geometry optimizations, transition state searches, and molecular dynamics simulations. The methodology is tested on the potential energy surfaces for separating ions and the dissociative phosphoryl transfer mechanism of methyl phosphate. The convergence behavior of the smooth COSMO method with respect to discretization level is examined and the numerical stability of the energy and gradient are compared to that from conventional COSMO calculations. The present method is further tested in applications to energy minimum and transition state geometry optimizations of neutral and charged metaphosphates, phosphates, and phosphoranes that are models for stationary points in transphosphorylation reaction pathways of enzymes and ribozymes. The results indicate that the smooth COSMO method greatly enhances the stability of quantum mechanical geometry optimization and transition state search calculations that would routinely fail with conventional solvation methods. The present MNDO/d-SCOSMO method has considerable computational advantages over hybrid quantum mechanical/molecular mechanical methods with explicit solvation, and represents a potentially useful tool in the arsenal of multi-scale quantum models used to study biochemical reactions. Read More View Full Article Download PDF |
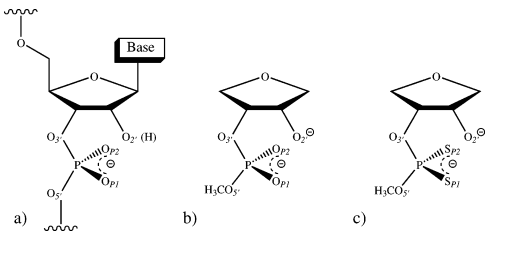 | Smooth solvation method for d-orbital semiempirical calculations of biological reactions. 2. Application to transphosphorylation thio effects in solution The Journal of Physical Chemistry B (2005) 109, 9810-9817 DOI: 10.1021/jp044061l 16852181Density-functional and semiempirical quantum methods and continuum dielectric and explicit solvation models are applied to study the role of solvation on the stabilization of native and thio-substituted transphosphorylation reactions. Extensive comparison is made between results obtained from the different methods. For the semiempirical methods, explicit solvation was treated using a hybrid quantum mechanical/molecular mechanical (QM/MM) approach and the implicit solvation was treated using a recently developed smooth solvation model implemented into a d-orbital semiempirical framework (MNDO/d-SCOSMO) within CHARMM. The different quantum and solvation methods were applied to the transesterification of a 3‘-ribose,5‘-methyl phosphodiester that serves as a nonenzymatic model for the self-cleavage reaction catalyzed by the hammerhead and hairpin ribozymes. Thio effects were studied for a double sulfur substitution at the nonbridging phosphoryl oxygen positions. The reaction profiles of both the native and double sulfur-substituted reactions from the MNDO/d-SCOSMO calculations were similar to the QM/MM results and consistent with the experimentally observed trends. These results underscore the need for a d-orbital semiempirical representation for phosphorus and sulfur for the study of experimentally observed thio effects in enzymatic and nonenzymatic phosphoryl transfer reactions. One of the major advantages of the present approach is that it can be applied to model chemical reactions at a significantly lower computational cost than either the density-functional calculations with implicit solvation or the semiempirical QM/MM simulations with explicit solvent. Read More View Full Article Download PDF |
 | High-order discretization schemes for biochemical applications of boundary element solvation and variational electrostatic projection methods The Journal of Chemical Physics (2005) 122, 194110 DOI: 10.1063/1.1899146 16161566A series of high-order surface element discretization schemes for variational boundary element methods are introduced. The surface elements are chosen in accord with angular quadrature rules for integration of spherical harmonics. Surface element interactions are modeled by Coulomb integrals between spherical Gaussian functions with exponents chosen to reproduce the exact variational energy and Gauss’s law for a point charge in a spherical cavity. The present work allows high-order surface element expansions to be made for variational methods such as the conductorlike screening model for solvation and the variational electrostatic projection method for generalized solvent boundary potentials in molecular simulations. Read More View Full Article Download PDF |
 | Pseudorotation barriers of biological oxyphosphoranes: a challenge for simulations of ribozyme catalysis Chemistry (2005) 11, 2081-2093 DOI: 10.1002/chem.200400790 15714539Pseudorotation reactions of biologically relevant oxyphosphoranes were studied by using density functional and continuum solvation methods. A series of 16 pseudorotation reactions involving acyclic and cyclic oxyphosphoranes in neutral and monoanionic (singly deprotonated) forms were studied, in addition to pseudorotation of PF5. The effect of solvent was treated by using three different solvation models for comparison. The barriers to pseudorotation ranged from 1.5 to 8.1 kcal mol-1 and were influenced systematically by charge state, apicophilicity of ligands, intramolecular hydrogen bonding, cyclic structure and solvation. Barriers to pseudorotation for monoanionic phosphoranes occur with the anionic oxo ligand as the pivotal atom, and are generally lower than for neutral phosphoranes. The OCH3 groups were observed to be more apicophilic than OH groups, and hence pseudorotations that involve axial OCH3/equatorial OH exchange had higher reaction and activation free energy values. Solvent generally lowered barriers relative to the gas-phase reactions. These results, together with isotope 18O exchange experiments, support the assertion that dianionic phosphoranes are not sufficiently long-lived to undergo pseudorotation. Comparison of the density functional results with those from several semiempirical quantum models highlight a challenge for new-generation hybrid quantum mechanical/molecular mechanical potentials for non-enzymatic and enzymatic phosphoryl transfer reactions: the reliable modeling of pseudorotation processes. Read More View Full Article Download PDF |
 | Ellipticity: a convenient tool to characterize electrocyclic reactions Chemistry (2005) 11, 1734-1738 DOI: 10.1002/chem.200401026 15669038The ellipticity of the electron density is proposed as a convenient tool to characterize electrocyclic reactions. The study of the electron density offers several advantages since it is an experimental observable. This simple topological index has proven powerful and robust for the characterization of a wide variety of electrocyclic and pseudoelectrocyclic processes. This property is sensitive to changes in the anisotropy of the electron density in the bond-forming region and provides an insightful description of the events occurring along the reaction coordinate. Read More View Full Article Download PDF |
 | The contribution of phosphate-phosphate repulsions to the free energy of DNA bending Nucleic Acids Research (2005) 33, 1257-1268 DOI: 10.1093/nar/gki272 PMC552960DNA bending is important for the packaging of genetic material, regulation of gene expression and interaction of nucleic acids with proteins. Consequently, it is of considerable interest to quantify the energetic factors that must be overcome to induce bending of DNA, such as base stacking and phosphate–phosphate repulsions. In the present work, the electrostatic contribution of phosphate–phosphate repulsions to the free energy of bending DNA is examined for 71 bp linear and bent-form model structures. The bent DNA model was based on the crystallographic structure of a full turn of DNA in a nucleosome core particle. A Green's function approach based on a linear-scaling smooth conductor-like screening model was applied to ascertain the contribution of individual phosphate–phosphate repulsions and overall electrostatic stabilization in aqueous solution. The effect of charge neutralization by site-bound ions was considered using Monte Carlo simulation to characterize the distribution of ion occupations and contribution of phosphate repulsions to the free energy of bending as a function of counterion load. The calculations predict that the phosphate–phosphate repulsions account for ~30% of the total free energy required to bend DNA from canonical linear B-form into the conformation found in the nucleosome core particle. Read More View Full Article Download PDF |
 | Variational Electrostatic Projection (VEP) methods for efficient modeling of the macromolecular electrostatic and solvation environment in activated dynamics simulations The Journal of Physical Chemistry B (2005) 109, 536-556 DOI: 10.1021/jp0469968 16851046New methods for the calculation of electrostatic interactions between the active dynamical region and surrounding external solvated macromolecular environment in hybrid quantum mechanical/molecular mechanical (QM/MM) simulations are presented. The variational electrostatic projection (VEP) method, and related variational reverse-mapping procedure (VEP-RVM) utilize an expansion in Gaussian surface elements for representation of electrostatic interactions. The use of a discretized surface that surrounds the active dynamical region greatly reduces the number of interactions with the particles of the external environment. The methods are tested on two catalytic RNA systems:? the hammerhead and the hairpin ribozymes. It is shown that with surface elements numbering from 302 to 1202 points the direct VEP and VEP-RVM methods are able to obtain relative force errors in the range of 0.5-0.05% and 0.09-0.0001%, respectively, using a 4.0 Å projection buffer. These results are encouraging and provide an essential step in the development of new variational macromolecular solvent-boundary methods for QM/MM calculations of enzyme reactions. Read More View Full Article Download PDF |
 | An Efficient Linear-Scaling Ewald Method for Long-Range Electrostatic Interactions in Combined QM/MM Calculations Journal of Chemical Theory and Computation (2005) 1, 2-13 DOI: 10.1021/ct049941i 26641110A method is presented for the efficient evaluation of long-range electrostatic forces in combined quantum mechanical and molecular mechanical (QM/MM) calculations of periodic systems. The QM/MM-Ewald method is a linear-scaling electrostatic method that utilizes the particle mesh Ewald algorithm for calculation of point charge interactions of molecular mechanical atoms and a real-space multipolar expansion for the quantum mechanical electrostatic terms plus a pairwise periodic correction factor for the QM and QM/MM interactions that does not need to be re-evaluated during the self-consistent field procedure. The method is tested in a series of molecular dynamics simulations of the ion-ion association of ammonium chloride and ammonium metaphosphate and the dissociative phosphoryl transfer of methyl phosphate and acetyl phosphate. Results from periodic boundary molecular dynamics (PBMD) simulations employing the QM/MM-Ewald method are compared with corresponding PBMD simulations using electrostatic cutoffs and with results from nonperiodic stochastic boundary molecular dynamics (SBMD) simulations, with cutoffs and with full electrostatics (no cutoff). The present method allows extension of linear-scaling Ewald methods to molecular simulations of enzyme and ribozyme reactions that use combined QM/MM potentials. Read More View Full Article Download PDF |
 | Theoretical methods that help understanding the structure and reactivity of gas phase ions International Journal of Mass Spectrometry (2005) 240, 37-99 DOI: 10.1016/j.ijms.2004.09.018 The methods of the quantum electronic structure theory are reviewed and their implementation for the gas phase chemistry emphasized. Ab initio molecular orbital theory, density functional theory, quantum Monte Carlo theory and the methods to calculate the rate of complex chemical reactions in the gas phase are considered. Relativistic effects, other than the spin–orbit coupling effects, have not been considered. Rather than write down the main equations without further comments on how they were obtained, we provide the reader with essentials of the background on which the theory has been developed and the equations derived. We committed ourselves to place equations in their own proper perspective, so that the reader can appreciate more profoundly the subtleties of the theory underlying the equations themselves. Finally, a number of examples that illustrate the application of the theory are presented and discussed. Read More View Full Article Download PDF |
 | Kinetic isotope effects on thio-substituted biological phosphoryl transfer reactions from density-functional theory Chemical Communications (2005) 31, 3909-3911 DOI: 10.1039/B502568K 16075068Primary and secondary kinetic and equilibrium isotope effects are calculated with density-functional methods for the dianionic methanolysis of the native (unsubstituted) and thio-substituted ethylene phosphates. Read More View Full Article Download PDF |
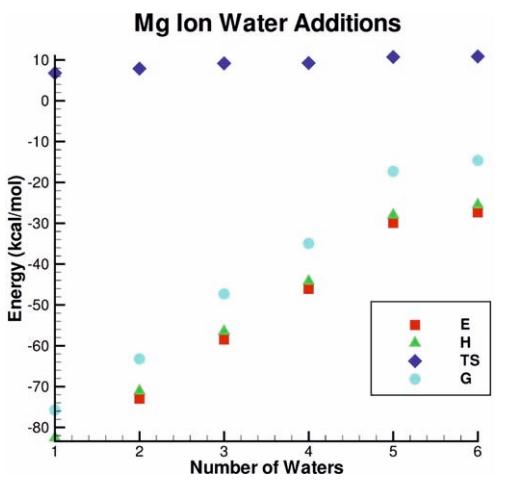 | Structure and binding of Mg(II) ions and di-metal bridge complexes with biological phosphates and phosphoranes Journal of Biological Inorganic Chemistry (2004) 9, 807-817 DOI: 10.1007/s00775-004-0608-2 15328556Divalent Mg2+ ions often serve as cofactors in enzyme or ribozyme-catalyzed phosphoryl transfer reactions. In this work, the interaction of Mg2+ ions and di-metal bridge complexes with phosphates, phosphoranes, and other biological ligands relevant to RNA catalysis are characterized with density functional methods. The effect of bulk solvent is treated with two continuum solvation methods (PCM and COSMO) for comparison. The relative binding affinity for different biological ligands to Mg2+ are quantified in different protonation states. The structure and stability of the single-metal and di-metal complexes are characterized, and the changes in phosphate and phosphorane geometry induced by metal ion binding are discussed. Di-metal bridge complexes are a ubiquitous motif and the key factors governing their electrostatic stabilization are outlined. The results presented here provide quantitative characterization of metal ion binding to ligands of importance to RNA catalysis, and lay the groundwork for design of new generation quantum models that can be applied to the full biological enzymatic systems. Read More View Full Article Download PDF |
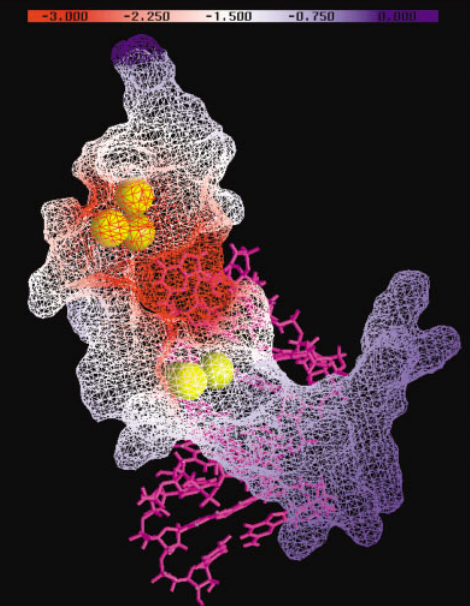 | Quantum descriptors for biological macromolecules from linear-scaling electronic structure methods Proteins: Structure, Function, and Bioinformatics (2004) 56, 724-737 DOI: 10.1002/prot.20171 15281126The characterization of electrostatic and chemical properties at the surface of biological macromolecules is of interest in elucidating the fundamental biological structure-function relationships as well as in problems of rational drug design. This paper presents a set of macromolecular quantum descriptors for the characterization of biological macromolecules in solution that can be obtained with modest computational cost from linear-scaling semi-empirical quantum/solvation methods. The descriptors discussed include: solvent-polarized electrostatic surface potential maps, equilibrated atomic charges, Fukui reactivity indices, approximate local hardness maps, and relative proton potentials. These properties are applied to study the conformational dependence of the electrostatic surface potential of the solvated phosphate-binding protein mutant (T141D), the regioselectivity of the zinc finger domains of HIV-1 nucleocapsid (NC) protein, and the order of pKa values of acidic residues in turkey ovomucoid third domain (OMTKY3) and of the zinc-binding residues in the carboxyl terminal zinc finger of NC. In all cases, insight beyond that obtainable from purely classical models is gained and can be used to rationalize the experimental observations. The macromolecular quantum descriptors presented here greatly extend the arsenal of tools for macromolecular characterization and offer promise in applications to modern structure-based drug design. Read More View Full Article Download PDF |
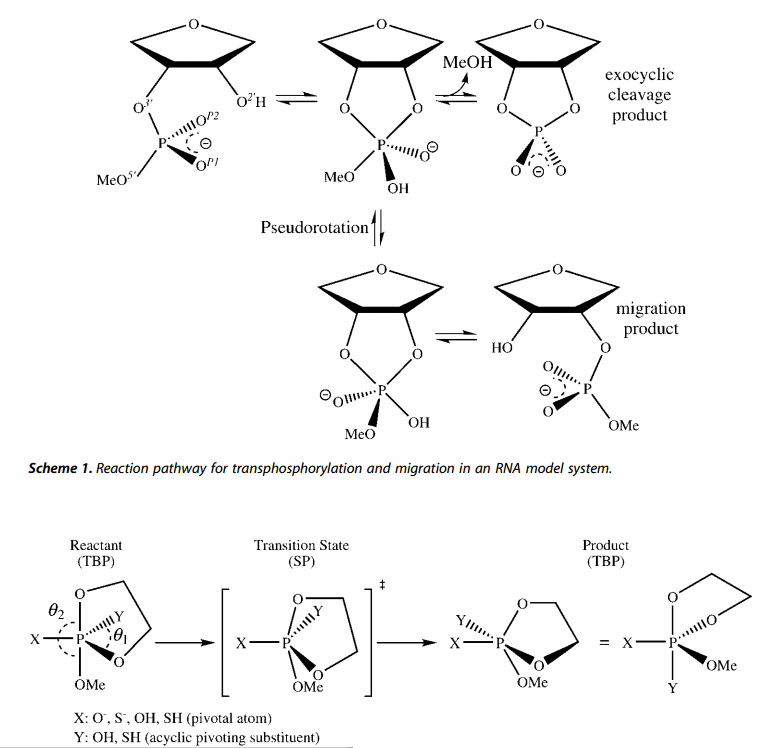 | Pseudorotation of natural and chemically modified biological phosphoranes: implications for RNA catalysis ChemPhysChem - A European Journal of Chemical Physics and Physical Chemistry (2004) 5, 1045-1049 DOI: 10.1002/cphc.200400091 15298394Model intermediates: Catalytic mechanisms of RNA transphosphorylation are investigated by monitoring changes in the reaction rate that occur upon substitution of key phosphate oxygen atoms with sulfur atoms. The mechanisms and barriers to pseudorotation for a series of oxyphosphorane and thiophosphorane molecules are presented (see picture). Theoretical models provide detailed insight into biological phosphorous reactivity in RNA systems. Read More View Full Article Download PDF |
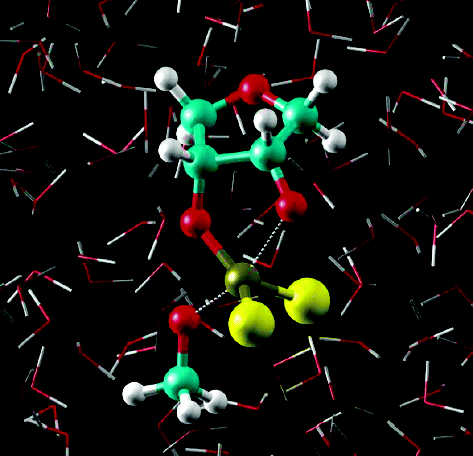 | Hybrid QM/MM study of thio effects in transphosphorylation reactions: the role of solvation Journal of the American Chemical Society (2004) 126, 7504-7513 DOI: 10.1021/ja031815l 15198597Transphosphorylation thio effects in solution are studied using hybrid QM/MM calculations with a d-orbital semiempirical Hamiltonian. Activated dynamics simulations were performed for a 3‘ ribose-phosphate model in an explicit 20 Å sphere of TIP3P water surrounded by a solvent boundary potential, and free energy analysis was performed using the weighted histogram analysis method. Single thio-substitutions at all of the phosphoryl oxygen positions and a double thio-substitution at the nonbridging positions were considered. The reaction free energy profiles are compared with available experimental data, and the role of solvation on the barrier heights and reaction coordinate is discussed. These results provide an important step in the characterization of thio effects in reactions of biological phosphates that may aid in the interpretation of kinetic data and ultimately help to unravel the catalytic mechanisms of ribozymes. Read More View Full Article Download PDF |
 | Many-body force field models based solely on pairwise Coulomb screening do not simultaneously reproduce correct gas-phase and condensed-phase polarizability limits The Journal of Chemical Physics (2004) 120, 9903-9906 DOI: 10.1063/1.1756583 15268007It is demonstrated that many-body force field models based solely on pairwise Coulomb screening cannot simultaneously reproduce both gas-phase and condensed-phase polarizability limits. Several many-body force field model forms are tested and compared with basis set-corrected ab initio results for a series of bifurcated water chains. Models are parameterized to reproduce the ab initio polarizability of an isolated water molecule, and pairwise damping functions are set to reproduce the polarizability of a water dimer as a function of dimer separation. When these models are applied to extended water chains, the polarization is over-predicted, and this over-polarization increased as a function of the overlap of molecular orbitals as the chains are compressed. This suggests that polarizable models based solely on pairwise Coulomb screening have some limitations, and that coupling with non-classical many-body effects, in particular exchange terms, may be important. Read More View Full Article Download PDF |
 | Theoretical Study of the Vinyl Allene Oxide to Cyclopent-2-en-1-one Rearrangement:? Mechanism, Torquoselectivity and Solvent Effects The Journal of Organic Chemistry (2004) 69, 3635-3644 DOI: 10.1021/jo049620z 15152991Density-functional calculations in the gas phase and solvent (PCM) at the B3LYP/6-311++G(3df,2p)//B3LYP/6-31++G(d,p) level were performed to study a series of six reactions that involve the rearrangement of vinyl allene oxides to cyclopent-2-en-1-ones along two distinct mechanistic pathways, namely concerted and stepwise. Calculations predict that stepwise pathways are highly competitive processes that occur via biradical/zwitterionic intermediates. Torquoselectivity is predicted to result from the concerted pathway leading to a stereodefined 4,5-disubstituted cyclopent-2-en-1-ones that should have memory of the starting terminal double-bond geometry and oxide configuration. The stepwise pathway cannot show torquoselectivity as cyclization of the planar oxidopentadienyl zwitterion can follow enantiomorphous conrotations. The concerted/stepwise mechanistic preference depends mainly on the olefin geometry and is further modulated by epoxide substitution. The influence of the solvent (PCM model for dichloromethane or water) is moderate, although the greater (de)stabilization of the polarized oxidopentadienyl zwitterions along the stepwise mechanism does alter the kinetic preferences exhibited by the systems in vacuo. Results with system 1e suggest that, if vinyl allene oxide II having a double bond with Z-geometry, an intermediate in the biogenesis of epi-jasmonic acid IV, is processed along an in stepwise mechanism following ring opening, the enzyme allene oxide cyclase must enforce enantiofacial torquoselectivity. Read More View Full Article Download PDF |
 | Complete basis set extrapolated potential energy, dipole, and polarizability surfaces of alkali halide ion-neutral weakly avoided crossings with and without applied electric fields The Journal of Chemical Physics (2004) 120, 7939 DOI: 10.1063/1.1690232 15267709Complete basis set extrapolations of alkali halide (LiF, LiCl, NaF, NaCl) energy, dipole, and polarizabilitysurfaces are performed with and without applied fields along the internuclear axis using state-averaged multireference configuration interaction. Comparison between properties (equilibrium separation, dissociation energy, crossing distance, diabatic coupling constant, dipole, and polarizability) derived from the extrapolated potential energy (or dipole) surfaces are made with those obtained from direct extrapolation from the basis set trends. The two extrapolation procedures are generally found to agree well for these systems. Crossing distances from this work are compared to those of previous work and values obtained from the Rittner potential. Complete basis set extrapolated crossing distances agree well with those derived from the Rittner potential for LiF, but were significantly larger for LiCl, NaF, and NaCl. The results presented here serve as an important set of benchmark data for the development of new-generation many-body force fields that are able to model charge transfer. Read More View Full Article Download PDF |
 | The structure and stability of biological metaphosphate, phosphate, and phosphorane compounds in the gas phase and in solution Journal of the American Chemical Society (2004) 126, 1654-1665 DOI: 10.1021/ja0356277 14871095Density functional calculations of a series of metaphosphates, acyclic and cyclic phosphates and phosphoranes relevant to RNA catalysis are presented. Solvent effects calculated with three well-established solvation models are analyzed and compared. The structure and stability of the compounds are characterized in terms of thermodynamic quantities for isomerization and ligand substitution reactions, gas-phase proton affinities, and microscopic solution pKa values. The large dataset of compounds allows the estimation of bond energies to determine the relative strengths of axial and equatorial P-O phosphorane single bonds and P-O single and double bonds in metaphosphates and phosphates. The relative apicophilicty of hydroxyl and methoxy ligands in phosphoranes are characterized. The results presented here provide quantitative insight into RNA catalysis and serve as a first step toward the construction of a high-level quantum database for development of new semiempirical Hamiltonian models for biological reactions. Read More View Full Article Download PDF |
 | Design and application of a multicoefficient correlatiomethod for dispersion interactions The Journal of Chemical Physics (2004) 120, 590 DOI: 10.1063/1.1630955 15267893A new multicoefficient correlation method (MCCM) is presented for the determination of accurate van der Waals interactions. The method utilizes a novel parametrization strategy that simultaneously fits to very high-level binding, Hartree–Fock and correlation energies of homo- and heteronuclear rare gas dimers of He, Ne, and Ar. The decomposition of the energy into Hartree–Fock and correlation components leads to a more transferable model. The method is applied to the krypton dimer system, rare gas–water interactions, and three-body interactions of rare gas trimers He3, Ne3, and Ar3. For the latter, a very high-level method that corrects the rare-gas two-body interactions to the total binding energy is introduced. A comparison with high-level CCSD(T) calculations using large basis sets demonstrates the MCCM method is transferable to a variety of systems not considered in the parametrization. The method allows dispersion interactions of larger systems to be studied reliably at a fraction of the computational cost, and offers a new tool for applications to rare-gas clusters, and the development of dispersion parameters for molecular simulation force fields and new semiempirical quantum models. Read More View Full Article Download PDF |
 | High-level ab initio methods for calculation of accurate potential energy surfaces of van der Waals complexes International Journal of Quantum Chemistry (2004) 98, 388-408 DOI: 10.1002/qua.20074 16178597A systematic series of highly correlated calculations of van der Waals potential energy surfaces (PESs) with large basis sets is presented. Reference data at the coupled-cluster theory restricted to single, double, and noniterative triple excitations [CCSD(T)] level with large singly augmented correlation-consistent basis functions, supplementary bond functions, and counterpoise corrections are provided. Results for minimum energy distances, well depths, vibrational frequencies, second virial coefficients, and Boyle temperatures are compared with corresponding experimental values. An extensive discussion of complete basis set extrapolation methods is presented. Here, the effect of extrapolation type, use of uncorrected and counterpoise-corrected PESs, and direct property extrapolation are analyzed. Last, a new multicoefficient correlation method for van der Waals potential energy surfaces (MCCM-vdW) is applied to three-body interactions of helium trimers and to He … H2O interactions. Comparison with high-level CCSD(T) calculations using large basis sets demonstrates that the MCCM-vdW method is transferable to systems not considered in its parameterization. The method allows dispersion interactions of much larger systems to be studied reliably at a fraction of computational cost and offers a new tool for applications to rare-gas clusters and the development of dispersion parameters for molecular simulation force fields. Read More View Full Article Download PDF |
 | Examination of the correlation energy and second virial coefficients from accurate ab initio calculations of rare-gas dimers The Journal of Chemical Physics (2003) 119, 2618-2622 DOI: 10.1063/1.1587684 Calculations of rare-gas dimers (He–He, Ne–Ne, Ar–Ar, He–Ne, He–Ar, and Ne–Ar) at the coupled-cluster single double (triple) level of theory with large basis sets including bond functions and counterpoise corrections are reported over a wide range of 100 internuclear separations. These results are compared to experimental curves obtained from fitting to rovibrational spectra, and to second virial coefficients and Boyle temperatures. Accurate analytic potentials are developed for the total interactionenergy, Hartree–Fock (exchange) energy, and correlation(dispersion)energy; the transferability of the latter is demonstrated to very high accuracy even in the region of considerable wave function overlap. These calculations represent an important set of benchmarks that can be used to develop improved empirical molecular mechanical force fields and new quantum models. Read More View Full Article Download PDF |
 | Insights into the regioselectivity and RNA-binding affinity of HIV-1 nucleocapsid protein from linear-scaling quantum methods Journal of Molecular Biology (2003) 330, 993-1004 DOI: 10.1016/S0022-2836(03)00658-2 12860122Human immunodeficiency virus type 1 (HIV-1) nucleocapsid protein (NC) plays several important roles in the viral life-cycle and presents an attractive target for rational drug design. Here, the macromolecular reactivity of NC and its binding to RNA is characterized through determination of electrostatic and chemical descriptors derived from linear-scaling quantum calculations in solution. The computational results offer a rationale for the experimentally observed susceptibility of the Cys49 thiolate toward small-molecule electrophilic agents, and support the recently proposed stepwise protonation mechanism of the C-terminal Zn-coordination complex. The distinctive binding mode of NC to SL2 and SL3 stem–loops of the HIV-1 genomic RNA packaging signal is studied on the basis of protein side-chain contributions to the electrostatic binding energies. These results indicate the importance of several basic residues in the 310 helical region and the N-terminal zinc finger, and rationalize the presence of several evolutionarily conserved residues in NC. The combined reactivity and RNA-binding study provides new insights that may contribute toward the structure-based design of anti-HIV therapies. Read More View Full Article Download PDF |
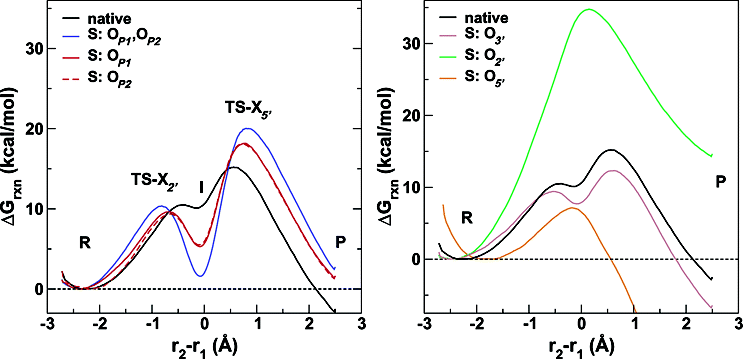 | Hybrid QM/MM study of thio effects in transphosphorylation reactions Journal of the American Chemical Society (2003) 125, 7178-7179 DOI: 10.1021/ja035167h 12797782Results of a series of hybrid quantum mechanical/molecular mechanical (QM/MM) activated dynamics simulations of thio effects in the transphosphorylation (methanolysis) of a 2‘-ribose, 5‘-methyl phosphate-diester under basic conditions are presented. Single and double substitutions in the nonbridging oxygen positions exhibit thio effects in accord with experimental data and show the existence of a stable intermediate. Thio substitution at the 2‘ and 5‘ positions resulted in reactions having a single transition state with increased and decreased free energy barriers, respectively, relative to the unsubstituted reaction. In all of the reactions except for the 5‘ substitution, the rate-limiting step corresponds to exocyclic cleavage. In the 5‘ substitution reaction, the rate-limiting step corresponds to endocyclic cleavage and shows a considerable reverse thio effect, in accord with experimental observations of phosphates with enhanced leaving groups. Thio substitution at the 3‘ position results in a mild reverse thio effect that arises from electronic stabilization of the dianionic transition state. The results presented here provide an important step toward the development and application of new hybrid QM/MM methods that, combined with experiment, may provide a detailed picture of the molecular mechanisms of RNA catalysis. Read More View Full Article Download PDF |
 | Parameterization of semiempirical methods to treat nucleophilic attacks to biological phosphates: AM1/d parameters for phosphorus Theoretical Chemistry Accounts (2003) 109, 149-159 DOI: 10.1007/s00214-002-0422-2 This paper reports a new AM1/d model for phosphorus that can be used to model nucleophilic attack of phosphates relevant for biological phosphate hydrolysis reactions. The parameters were derived from a quantum dataset calculated with hybrid density-functional theory [B3LYP/6-311++G(3df,2p)//B3LYP/6-31++G(d,p)] of phosphates and phosphoranes in various charge states, and on transitions states for nucleophilic attacks. A suite of non-linear optimization methods is outlined for semiempirical parameter development based on integrated evolutionary (genetic), Monte Carlo simulated annealing and direction set minimization algorithms. The performance of the new AM1/d model and the standard AM1 and MNDO/d models are compared with the density-functional results. The results demonstrate that the strategy of developing semiempirical parameters specific for biological reactions offers considerable promise for application to large-scale biological problems. Read More View Full Article Download PDF |
 | Preface to proceedings of the symposium on methods and applications of combined quantum mechanical and molecularmechanical potentials (2001) Theoretical Chemistry Accounts (2003) 109, 99 DOI: 10.1007/s00214-002-0414-2 Preface to proceedings of the symposium on methods and applications of combined quantum mechanical and molecularmechanical potentials (2001) Read More View Full Article Download PDF |
 | Fast approximate methods for calculating nucleic acid base pair interaction energies Journal of Computational Chemistry (2003) 24, 57-67 DOI: 10.1002/jcc.10150 12483675Interaction enthalpies for six base pairs have been computed at a variety of efficient levels of electronic structure theory and compared to experiment. In addition to previously defined levels of theory, modified Hamiltonians with adjusted parameters in hybrid Hartree–Fock/density functionals and semiempirical neglect-of-diatomic-differential-overlap models were examined. Of the pure and hybrid density functional levels, mPWPW91/MIDI! performed most satisfactorily, as judged by comparison not only to the available experimental data, but also to data from more robust electronic structure methods for 22 additional base pairs. The low computational cost of the mPWPW91/MIDI! model was further exploited in an investigation of various base trimers, tetramers, and one base pentamer. A carefully reparameterized semiempirical model, PM3BP, was able to achieve similar levels of accuracy at a still greater savings in terms of computational effort. Read More View Full Article Download PDF |
 | Time-dependent density functional theory calculations of molecular static and dynamic polarizabilities, cauchy coefficients and their anisotropies with atomic numerical basis functions Journal of Molecular Structure: THEOCHEM (2002) 591, 255-266 DOI: 10.1016/S0166-1280(02)00246-4 Static and dynamic polarizabilities of a range of small first row compounds have been calculated with time-dependent density functional theory in the local spin-density approximation using numerical atomic basis sets. The results are compared to earlier computational work, in particular the work of Van Caillie and Amos [C. Van Caillie, R.D. Amos, Chem. Phys. Lett. 291 (1998) 71], as well as experimental values. The results for static isotropic and anisotropic polarizabilities of H2O, N2, CO, NH3, and CH4 are in good agreement with previous calculations. The results for the dynamic polarizabilities as expressed in the S(-4) Cauchy coefficients and their anisotropies for H2O, N2, CO2, NH3, CH4, C2H2, C2H4 and C2H6 are also in good agreement with previous results. We have also explored the scaling of our implementation of the time-dependent coupled-perturbed Kohn–Sham equations by evaluating the static and dynamic polarizabilities of bifurcated water chains ranging from 1–20 molecules in size. Read More View Full Article Download PDF |
 | Quantum Mechanical Characterization of Nucleic Acids in Solution:? A Linear-Scaling Study of Charge Fluctuations in DNA and RNA The Journal of Physical Chemistry B (2002) 106, 7693-7703 DOI: 10.1021/jp0146667 Atom-centered point charges are a convenient and computationally efficient way to approximately represent the electrostatic properties of biological macromolecules. Atomic charges are routinely used in molecular modeling applications such as molecular simulations, molecular recognition, and ligand binding studies and for determining quantitative structure activity relationships. In the present paper a divide-and-conquer linear-scaling semiempirical method combined with a conductor-like screening model is applied to the calculation of charge distributions of solvated DNA and RNA duplexes in canonical A- and B-forms. The atomic charges on A-DNA, B-DNA, and A-RNA duplex decamers are analyzed to characterize the convergence of the linear-scaling method, and the effects of the charge model and semiempirical Hamiltonian. Furthermore, the inter- and intramolecular charge variations on DNA and RNA duplex 72-mers are investigated to gain insight into the influence of conformation, base stacking, and solvent polarization on the charge distributions. The charges derived from the linear-scaling semiempirical calculations reflect the electronic relaxation in the solvated macromolecular environment and therefore provide a better reference charge state for biomolecular modeling applications. Read More View Full Article Download PDF |
 | Spectroscopic Properties of Tyrosyl Radicals in Dipeptides Journal of the American Chemical Society (2002) 124, 5496-5505 DOI: 10.1021/ja0164327 11996592Redox-active tyrosine residues play important roles in long-distance electron reactions in enzymes, including prostaglandin H synthase, galactose oxidase, ribonucleotide reductase, and photosystem II. Magnetic resonance and vibrational spectroscopy provide methods with which to study the structures of redox-active amino acids in proteins. In this report, ultraviolet photolysis was used to generate tyrosyl radicals from polycrystalline tyrosinate or dipeptides, and the structure of the radical was investigated with EPR and reaction-induced FT-IR spectroscopy at 77 K. Photolysis at 77 K is expected to generate a neutral tyrosyl radical through oxidation of the aromatic ring. EPR and FT-IR results obtained from 13C-labeled tyrosine were consistent with that expectation. Surprisingly, labeling of the tyrosyl amino group with 15N also resulted in isotope-shifted bands in the photolysis spectrum. The force constant of a NH deformation mode increased when the tyrosyl radical was generated. These data suggest an interaction between the pi system of the tyrosyl radical and the amino group. In spectra acquired from the dipeptides, evidence for a sequence-dependent interaction between the tyrosyl radical and the amide bond of the dipeptide was also obtained. We postulate that perturbation of the amino or the amide/imide groups may occur through a spin polarization mechanism, which is indirectly detected as a change in NH force constant. This conclusion is supported by density functional calculations, which suggest a conformationally sensitive delocalization of spin density onto the amino and carboxylate groups of the tyrosyl radical. These experiments provide a step toward a detailed spectral interpretation for protein-based tyrosyl radicals. Read More View Full Article Download PDF |
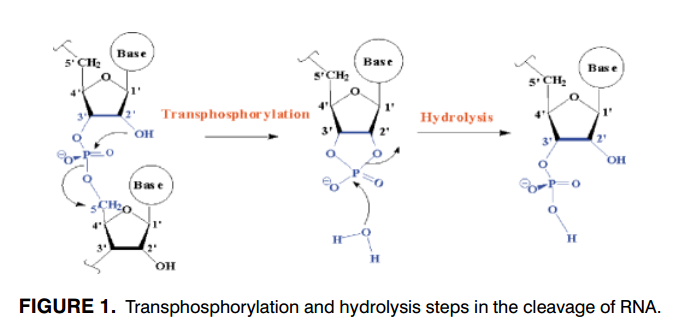 | Theoretical studies on the hydrolysis of phosphate diesters in the gas phase, solution, and RNase A International Journal of Quantum Chemistry (2002) 86, 10-26 DOI: 10.1002/qua.1601 Density functional theory, polarizable continuum models and semiempirical hybrid quantum mechanical/molecular mechanical (QM/MM) calculations were applied to the hydrolysis of phosphate diesters in the gas phase, in solution, and in the enzyme RNase A. Neutralization of the negative charge of the pentacovalent phosphorane intermediates provides a substantial stabilization of the transition-state structures in the gas phase. Inclusion of solvent effects on the phosphate/phosphorane species was critical to reproducing the trends in reactivity observed experimentally. Finally, the catalytic mechanism for the hydrolysis of uridine 2',3'-cyclic phosphate by RNase A was studied by QM/MM calculations. Our results suggest that the rate-limiting transition state of the reaction corresponds to the approach of a water molecule to the phosphate and its activation by His119. Thus, His119 acts as a generalized base for the reaction. The water attack leads to a pentacovalent phosphorane transition state of formal charge -2; this excess of negative charge in the transition state is stabilized by a number of positively charged residues including His12 and Lys41. In the second stage of the reaction, the phosphorane is converted into products. This part of the reaction proceeds without a detectable barrier, and it is facilitated by a proton transfer from Lys41 to the departing O2'. Read More View Full Article Download PDF |
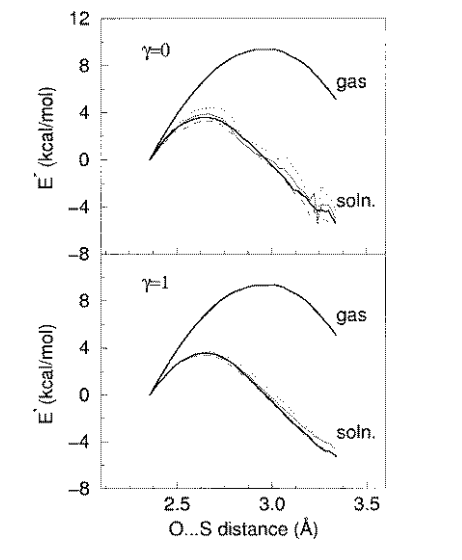 | Electronic structure properties of solvated biomolecules: a quantum approach for macromolecular characterization Journal of Computational Chemistry (2000) 21, 1562-1571 DOI: 10.1002/1096-987X(200012)21:16<1562::AID-JCC13>3.0.CO;2-E Linear-scaling electronic structure calculations of solvated biomolecules have been carried out using a semiempirical Hamiltonian and a new smooth solvation potential. These methods afford a new way of generating macromolecular properties that include quantum electronic structure. In addition to the widely used classical electrostatic potential maps based on empirically derived static point charges, now fully quantum mechanical electrostatic potentials that include electronic polarization are possible. Linear-scaling electronic structure methods provide a host of response properties of the electron density such as linear response functions, local hardness functions, and Fukui functions. It is the hope that these indices will extend insight into problems of biological macromolecular characterization. Read More View Full Article Download PDF |
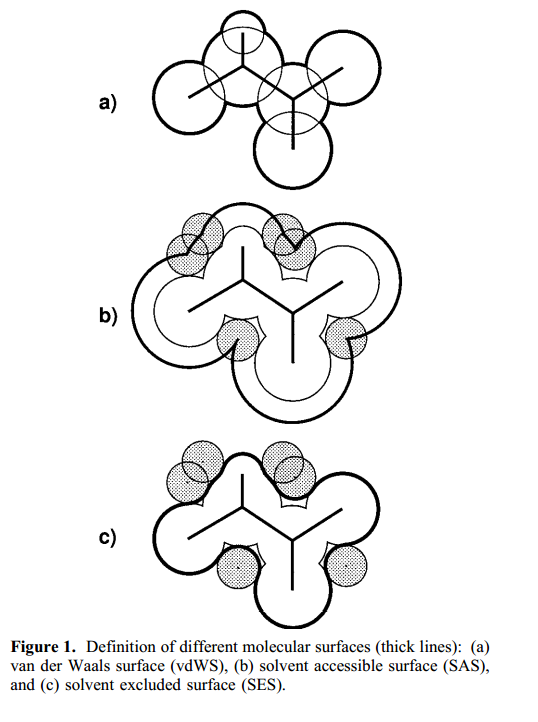 | A Smooth Solvation Potential Based on the Conductor-Like Screening Model The Journal of Physical Chemistry A (1999) 103, 11060-11079 DOI: 10.1021/jp992097l The development of a smooth solvation potential from which analytic derivatives can be derived is important for molecular applications that require geometry optimization and conformational sampling. Derivatives in conventional boundary element solvation methods are typically treated approximately, and contain singularities that arise from discontinuities in the potential. We present a simple smooth solvation potential that is based on the conductor-like screening model proposed by Klamt and Schüürmann (Klamt, A.; Schüürmann, G. J. Chem. Soc., Perkin. Trans. 2, 1993, 799). The model uses a simple solvent accessible surface with an atomic sphere discretization based on high-order angular quadrature schemes for spherical harmonics. Surface elements are modeled by spherical Gaussian functions with exponents calibrated to obtain the exact Born ion energy and uniform surface charge density and to avoid Coulomb singularities present in conventional point-charge surface element models. The set of linear equations are modified to produce a rigorously smooth solvation potential by allowing the effect of new surface elements to be turned on or off over a finite switching region around each atom. Numerical tests of the method are provided, in addition to discussions of rotational variance, generalization to arbitrary internal dielectric, use of constraints, and extension to a smooth surface area model. Read More View Full Article Download PDF |
 | Application of Linear-Scaling Electronic Structure Methods to the Study of Polarization of Proteins and DNA in Solution ACS Symposium Series (1998) 712, 275-287 DOI: 10.1021/bk-1998-0712.ch018 Semiempirical quantum calculations of biomolecular systems in solution were performed using recently developed linear-scaling methods to examine the role of solute polarization in the process of solvation. The solvation free energy of several protein and DNA molecules and complexes were computed and decomposed to asses the relative magnitude of electrostatic and polarization contributions. The effect of solvation and complex formation on the electronic density of states was also studied. Read More View Full Article Download PDF |
 | Quantum Mechanical Treatment of Biological Macromolecules in Solution using Linear-Scaling Electronic Structure Methods Physical Review Letters (1998) 80, 5011-5014 DOI: 10.1103/PhysRevLett.80.5011 A linear-scaling self-consistent field method for calculation of the electronic structure of biological macromolecules in solution is presented. The method is applied at the semiempirical Hartree-Fock level to the determination of heats of formation, solvation free energies, and density of electronic states for several protein and DNA systems. Read More View Full Article Download PDF |
 | Parameterization and efficient implementation of a solvent model for linear-scaling semiempirical quantum mechanical calculations of biological macromolecules Chemical Physics Letters (1996) 263, 297-304 DOI: 10.1016/S0009-2614(96)01198-0 A method is developed to include solvation effects in linear-scaling semiempirical quantum calculations. Favorable scaling of computational effort for large molecules is achieved using a preconditioned conjugate gradient technique in conjunction with a linear-scaling recursive bisection method for evaluation of electrostatic interactions. The method requires approximately 30% computational overhead relative to gas-phase calculations. Effective atomic radii for biological macromolecules are derived from fitting to experimental and theoretical solvation energies for small molecules homologous to amino- and nucleic acid residues. Read More View Full Article Download PDF |
 | Density-Functional Study of the Geometries, Stabilities, and Bond Energies of Group III-V (13-15) Four-Membered-Ring Compounds Journal of the American Chemical Society (1996) 118, 5732-5736 DOI: 10.1021/ja951706+ A theoretical investigation has been carried out on several group III-V (13-15) four-membered-ring compounds which, if experimentally attainable, are potentially useful as precursors to nanocrystalline electronic and semiconductor materials. Four-membered-ring compounds considered in this study have core structures of the following form:? MEM‘E‘ and MEMX (M, M‘ = In, Ga, Al; E, E‘ = P, As; X = Cl, Br). Equilibrium geometries, binding energies, and bond energies were determined based on local density approximation (LDA) and gradient-corrected density-functional methods. Optimized ring geometries obtained with LDA agree closely with single-crystal X-ray crystallographic structures of known compounds with the same four-membered-ring cores. The following trends in bond energies are observed:? M-Cl » M-P > M-As » M-Br (M = In, Ga, Al), and Al-Y > Ga-Y > In-Y (Y = P, As, Cl, Br). Although only one M-Br-containing mixed-bridge four-membered-ring compound has been reported and no such Al-Cl-containing mixed-bridge species have yet been synthesized, our calculations suggest that compounds containing these two ring systems are stable. Read More View Full Article Download PDF |
 | Quantum Mechanical Study of Aqueous Polarization Effects on Biological Macromolecules Journal of the American Chemical Society (1996) 118, 10940-10941 DOI: 10.1021/ja961937w Methods to determine the electronic structure of atoms and molecules are crucial for a reliable description of complex chemical processes that are inaccessible to conventional empirical models. Standard quantum mechanical techniques are limited to fairly small molecular systems due to unfavorable scaling properties inherent in the computational algorithms. We report the application of a recently developed linear-scaling quantum mechanical method to the study of aqueous polarization effects on biological macromolecules. The polarization contribution to the solvation free energy is in the range of 5 - 15% for proteins and 1 - 3% for DNA. Results suggest that polarization of proteins and DNA in the process of solvation can be well approximated by a linear response model. The developments presented here extend the realm of quantum chemical techniques to applications of macromolecular systems in solution. Read More View Full Article Download PDF |
 | Linear-scaling semiempirical quantum calculations for macromolecules The Journal of Chemical Physics (1996) 105, 2744 DOI: 10.1063/1.472136 A linear-scaling method to carry out semiempirical quantum mechanical calculations for large systems has been developed based on the density matrix version of the divide-and-conquer approach. The method has been tested and demonstrated to be accurate and efficient. With this implementation, semiempirical quantum mechanical calculations are made possible for large molecules over 9000 atoms on a typical workstation. For biological macromolecules,solvent effects are included with a dielectric continuum model. Read More View Full Article Download PDF |
 | A chemical potential equalization method for molecular simulations The Journal of Chemical Physics (1996) 104, 159-172 DOI: 10.1063/1.470886 A formulation of the chemical potential (electronegativity) equalization principle is presented from the perspective of density-functional theory. The resulting equations provide a linear-response framework for describing the redistribution of electrons upon perturbation by an applied field. The method has two main advantages over existing electronegativity equalization and charge equilibration methods that allow extension to accurate molecular dynamics simulations. Firstly, the expansion of the energy is taken about the molecular ground state instead of the neutral atom ground states; hence, in the absence of an external field, the molecular charge distribution can be represented by static point charges and dipoles obtained from fitting to high-level ab initio calculations without modification. Secondly, in the presence of applied fields or interactions with other molecules, the density response can be modeled accurately using basis functions. Inclusion of basis functions with dipolar or higher order multipolar character allows molecules or chemical groups to have correct local anisotropicpolarizabilities. A modified semiempirical form of the hardness matrix has been introduced that can be evaluated efficiently using Gaussians, and requires only one parameter per basis function. Applications at two basis-set levels demonstrate the method can accurately reproduce induced dipole moments and estimated chemical potentials obtained from density-functional calculations for a variety of molecules. Inclusion of basis functions beyond the conventional spherical-atom type is essential in some instances. The present formulation provides the foundation for a promising semi-empirical model for polarization and charge transfer in molecular simulations. Read More View Full Article Download PDF |
 | A new definition of atomic charges based on a variational principle for the electrostatic potential energy The Journal of Chemical Physics (1995) 102, 7549 DOI: 10.1063/1.469086 A unique definition of atomic charges in molecules is presented based on a variational principle involving the electrostatic potential energy. The method requires only the electron density as input, and does not rely on an arbitrary set of fitting points as do conventional electrostatic potential fitting procedures. The dipole moments and electrostatic potentials calculated from atomic charges obtained from this method agree well with those from self-consistent-field calculations. The new method also provides a spherical-atom potential model that may be useful in future generation molecular simulation force fields. Read More View Full Article Download PDF |
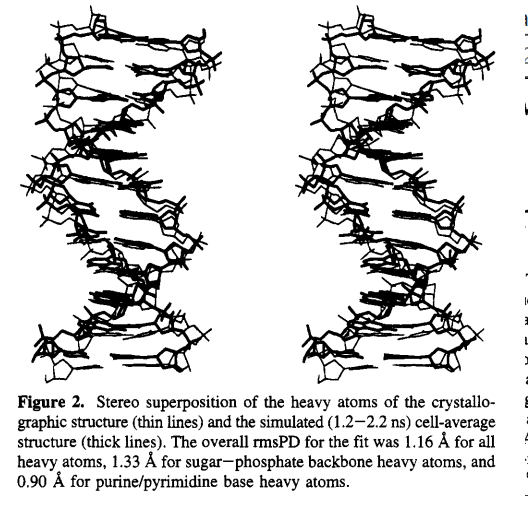 | Toward the Accurate Modeling of DNA: The Importance of Long-Range Electrostatics Journal of the American Chemical Society (1995) 117, 5001-5002 DOI: 10.1021/ja00122a034 The relationship between DNA structure and function is fundamental to the understanding of biological processes. Currently, the most reliable source of biomolecular structural information comes from X-ray crystallographic data. Recent advances in theoretical modeling techniques have allowed molecular simulations to approach the accuracy obtainable from X-ray crystallography for proteins. We report the results of a 2.2 ns simulation of the B-DNA dodecamer d[CGCGAATTCGCG12 in a crystal unit cell, and demonstrate that, with rigorous accommodation of long-range forces, molecular simulation may be extended to provide atomic level accuracy of polynucleotide structures. Read More View Full Article Download PDF |
 | The effect of hydrostatic pressure on protein crystals investigated by molecular simulation The Jerusalem Symposia on Quantum Chemistry and Biochemistry (1995) 27, 203-215 DOI: 10.1007/978-94-011-0497-5_17 The effect of hydrostatic pressure on protein crystal structures is examined with molecular simulation. Four 1 ns molecular dynamics simulations of bovine pancreatic trypsin inhibitor in a crystal unit cell have been performed at solvent densities corresponding to 32%, 36%, 40%, and 44% solvent. Electrostatic interactions in the crystalline environment were treated rigorously with Ewald sums. The effect of varying the solvent density at constant unit cell volume is analyzed with respect to changes in protein structure, atomic fluctuations, solvation, and crystal packing. The results indicate the solvent density range 36–40% gives excellent overall agreement with high resolution crystallographic data (~O.3Å rms backbone deviation). The low density (32%) and high density (44%) simulations have larger deviations. Read More View Full Article Download PDF |
 | A generalized formulation of electronegativity equalization from density-functional theory International Journal of Quantum Chemistry (1995) 56, 385-394 DOI: 10.1002/qua.560560842 A generalized formulation of the electronegativity equalization principle is presented from the perspective of density-functional theory. The resulting equations provide a linear-response framework for describing the redistribution of electrons upon perturbation by an external density or applied field. The equations can be solved using a finite set of basis functions to model the density response. Applications demonstrate the method accurately reproduces dipole moments and chemical potentials obtained from density-functional calculations. The method provides high accuracy in the presence of relatively strong perturbations such as those arising from interactions with other molecules or applied fields, and is “exact” in the limit that these interactions vanish. The method has the advantage that accuracy can be systematically improved by inclusion of more complete basis functions. The present formulation provides the foundation for a promising semiempirical model for polarization and charge transfer in molecular simulations. Read More View Full Article Download PDF |
 | The Fast Fourier Poisson Method for Calculating Ewald Sums The Journal of Chemical Physics (1994) 101, 3298 DOI: 10.1063/1.467576 The conventional Ewald expression for the electrostatic energy and forces is recast in a form that can be evaluated to high accuracy in order N?log(N) steps using fast Fourier transforms. The fast Fourier Poisson method does not rely on interpolation approaches or Taylor/multipole expansions, and can be easily integrated with conventional molecular dynamics algorithms. Read More View Full Article Download PDF |
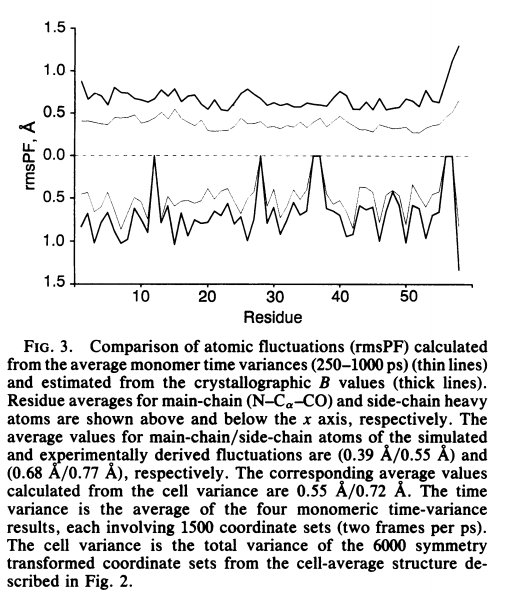 | Atomic-level Accuracy in Simulations of Large Protein Crystals Proceedings of the National Academy of Sciences of the United States of America (1994) 91, 8715-8718 DOI: 10.1073/pnas.91.18.8715 PMC44677Proper treatment of long-range Coulombic forces presents a major obstacle to providing realistic molecular dynamics simulations of macromolecules. Traditional approximations made to lessen computational cost ultimately lead to unrealistic behavior. The particle mesh Ewald method accommodates long-range Coulombic forces accurately and efficiently by use of fast Fourier transform techniques. We report a 1-ns simulation of bovine pancreatic trypsin inhibitor in a crystal unit cell using the particle mesh Ewald methodology. We find an rms backbone deviation from the x-ray structure (0.33 A) that is lower than that observed between bovine pancreatic trypsin inhibitor in different crystal forms and much lower than those of previous simulations. These results bridge the gap between structures obtained from molecular simulation and those from experiment. Read More View Full Article Download PDF |
 | Density-functional Calculations of the Structure and Stability of C240 Physical Review B: Condensed Matter and Materials Physics (1994) 49, 8526-8528 DOI: 10.1103/PhysRevB.49.8526 Density-functional calculations have been performed to determine optimized geometries and energies of C240 using the divide-and-conquer method. Six initial geometries were considered, resulting in convergence to two optimized configurations. The formation energies of the optimized structures are separated by approximately 0.07 eV/carbon atom. The lower-energy structure is highly spherical in agreement with preliminary studies and experimental observations. The higher-energy structure is polyhedrally faceted. The results support the conclusion that the most stable form of large carbon clusters is that of dense spherical caged structures. Read More View Full Article Download PDF |
 | Simulations of the Solution Structure of the HIV-1 Protease in the Presence and Absence of Bound Zinc Journal of Computational Chemistry (1994) 15, 61-71 DOI: 10.1002/jcc.540150108 Zinc ions have been shown to inhibit human immunodeficiency virus type 1 (HIV-1) protease in vitro at neutral pH [Zhang et al. Biochemistry, 36, 8717 (1991)]. Kinetic data from this study support a reversible binding mechanism of zinc in the active site. Preliminary calculations of the ion–protein potential energy based on the geometry of the crystallographic structure [Wlodawer et al. Science, 245, 616 (1989)] are consistent with this proposed mechanism. To examine the structure of HIV-1 protease with zinc bound in the active site, molecular dynamics simulations in the presence and absence of zinc at this site have been carried out to 200 ps. These simulations suggest zinc remains stably bound to the catalytic aspartate residues without disruption of the dimer or significant alteration of the active site structure. These data are consistent with those observed by Zhang et al. (1991), and together give strong evidence that this is the binding site that leads to inactivation. A proposed model of zinc binding at the active site based on quantum mechanical calculations indicates Zn+2 coordination is monodentate with each catalytic aspartate, leaving at least two ligand positions potentially free (occupied by water molecules in the calculations). Read More View Full Article Download PDF |
 | The Effect of Long-range Electrostatic Interactions in Simulations of Macromolecular Crystals: A Comparison of the Ewald and Truncated List Methods The Journal of Chemical Physics (1993) 99, 8345 DOI: 10.1063/1.465608 Simulations of the HIV-1 protease unit cell using a 9 Å cutoff, 9/18 Å ‘‘twin-range’’ cutoff, and full Ewald sums have been carried out to 300 ps. The results indicate that long-range electrostaticinteractions are essential for proper representation of the HIV-1 protease crystal structure. The 9 Å simulation did not converge in 300 ps. Inclusion of a 9/18 Å ‘‘twin-range’’ cutoff showed significant improvement. Simulation using the Ewald summation convention gave the best overall agreement with x-ray crystallographic data, and showed the least internal differences in the time average structures of the asymmetric units. The Ewald simulation represents an efficient implementation of the Particle Mesh Ewald method [Darden et al., J. Chem. Phys. 98, 10?089 (1993)], and illustrates the importance of including long-range electrostatic forces in large macromolecular systems. Read More View Full Article Download PDF |
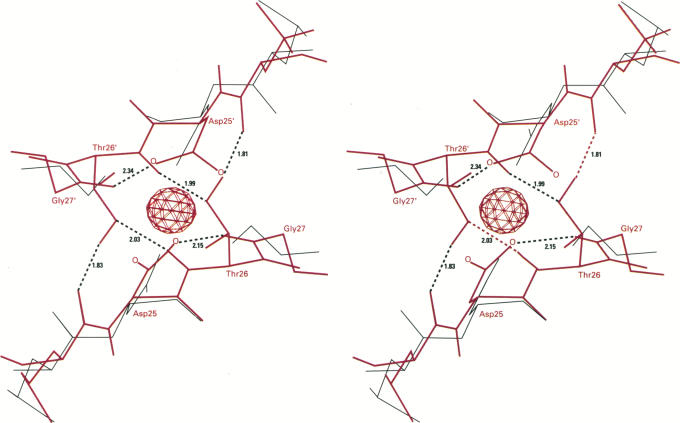 | Molecular Modeling Studies Suggest that Zinc Ions Inhibit HIV-1 Protease by Binding at Catalytic Aspartates Environmental Health Perspectives (1993) 101, 246-250 DOI: 10.1289/ehp.93101246 PMC1519788Human immunodeficiency virus type 1 protease is inhibited in vitro by zinc ions at neutral pH. The binding site of these ions is not known; however, experimental data suggest that binding may occur in the active site. To examine the possibility of zinc binding in the active site, molecular dynamics simulations in the presence and absence of zinc have been carried out to 200 psec. The results are compared with the 2.8-A crystallographic structures of a synthetic HIV-1 protease, and a zinc binding site at the catalytic aspartate residues (Asp-25, Asp-25') is proposed. Molecular dynamics simulations show that the zinc ion remains stably bound in this region, coordinating the carboxylate side chains of both aspartate residues. Interaction with zinc does not disrupt the dimeric structure of the protein or significantly alter the structure of the active site. These data are consistent with experimental studies of HIV-1 protease inhibition by zinc and give strong evidence that this is the binding site that leads to inactivation. Read More View Full Article Download PDF |
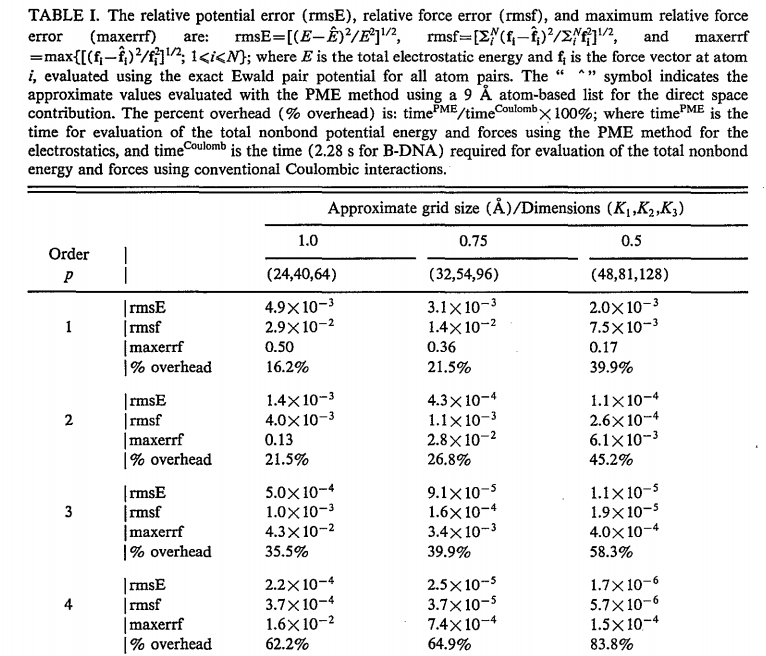 | Particle Mesh Ewald: An N·log(N) Method for Ewald Sums in Large Systems The Journal of Chemical Physics (1993) 98, 10089 DOI: 10.1063/1.464397 An N·log(N) method for evaluating electrostatic energies and forces of large periodic systems is presented. The method is based on interpolation of the reciprocal space Ewald sums and evaluation of the resulting convolutions using fast Fourier transforms. Timings and accuracies are presented for three large crystalline ionic systems. Read More View Full Article Download PDF |
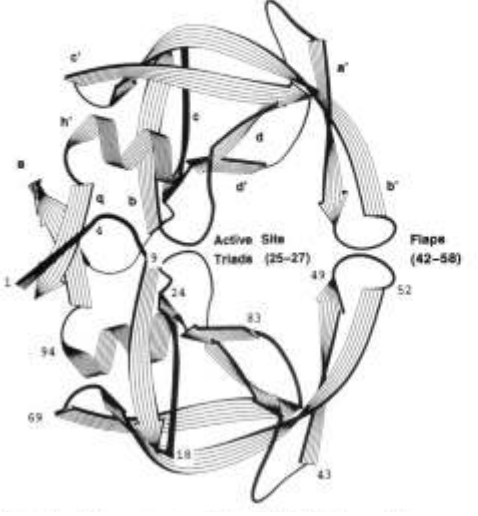 | Molecular Dynamics Simulation of HIV-1 Protease in a Crystalline Environment and in Solution Biochemistry (1993) 32, 1443-1453 DOI: 10.1021/bi00057a007 8431424Simulations of the unbound form of the human immunodeficiency virus type 1 protease have been carried out to 200 ps in a crystalline environment and in solution. Solution simulations were performed with and without charge-balancing counterions. The results are compared with the 2.8-A crystallographic structure of Wlodawer et al. [(1989) Science 245, 6161, and a proposed model for the solution structure which involves local refolding of the flap regions is presented. The simulations suggest the crystal packing environment of the protease dimer stabilizes the flaps in an extended conformation. Solvation of the dimer leads to local refolding of the flaps which contract toward the active site, forming increased overlap and stronger intersubunit hydrogn bonding at the tips. The degree to which the flaps overlap in solution is observed to depend on the charge state of the system. Read More View Full Article Download PDF |
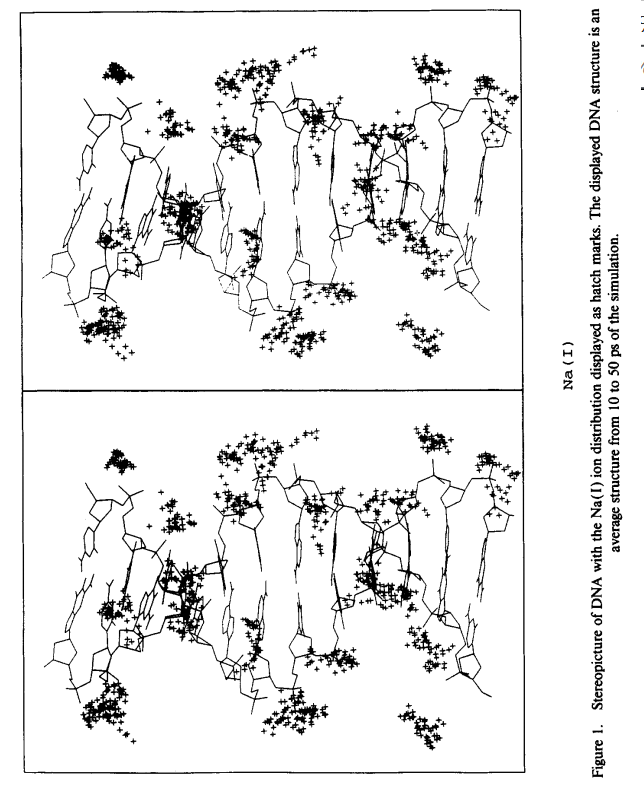 | The Interaction of Na(I), Ca(II), and Mg(II) Metal Ions with Duplex DNA: A Theoretical Modeling Study International Journal of Quantum Chemistry (1992) 44, 145-166 DOI: 10.1002/qua.560440715 DNA is a negatively charged biopolymer composed of monomeric nucleotides each carrying on average a net (-1) charge concentrated in the region of the phosphate backbone. In solution, the net negative charge of the molecule is presumably balanced by positively charged cations that interact with the DNA. Important questions arise as to the molecular details of the interaction of different cations with DNA. In this paper, we investigate the interaction of monovalent sodium ions and divalent magnesium and calcium ions with duplex DNA using molecular dynamics. Three 50 ps molecular dynamics simulations of the DNA sequence d[CGCGAATTCGCG]2 have been performed in different ionic environments. Each system is constructed to be electrically neutral and is composed of the DNA duplex immersed in a large water bath containing a specific ionic species [Na(I), Mg(II), or Ca(II)]. The structural differences of the DNA itself as a result of interacting with each ionic species have been previously examined (D. M. York, T. Darden, D. Deerfield, II, and L. Pedersen, J. Biomol. Struct. Dyn., submitted). In the current work, the ion–DNA and ion–water interactions are examined. The coordination shells of the ions and distributions around the phosphate anions are reported, and both show close encouraging agreement with available experimental work. The results of our studies indicate that the hydration state of the ion plays an important role in the direct coordination of the phosphate anions. Na(I) and Ca(II) ions prefer to coordinate the phosphate anions directly, whereas Mg(II) ions have a greater tendency to interact with phosphate anions as fully hydrated cations. Read More View Full Article Download PDF |