
Zeke Piskulich

 Position: Postdoctoral Associate
Position: Postdoctoral AssociateOffice: CIPR-308
About Me:
My background is in chemical dynamics, biophysics, spectroscopy, and multi-scale modeling of biochemical systems from the very small (Quantum Mechanics/Molecular Mechanics) to the very large (Coarse-Grained Molecular Dynamics). I'm currently using a variety of tools, such as Alchemical Free Energy Calculations, to understand small molecule binding to nucleic acids.
Full Publications: | |
|---|---|
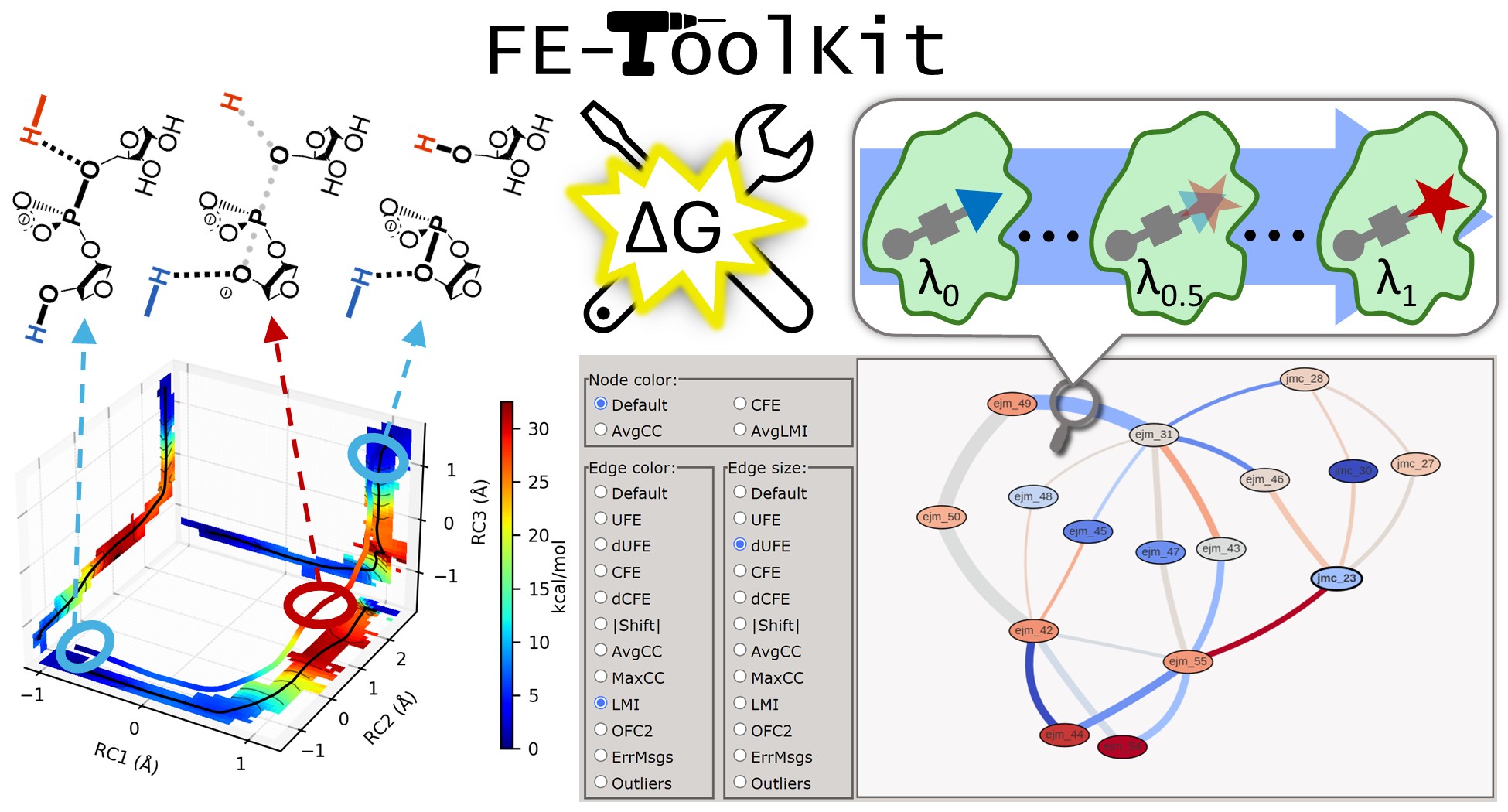 | FE-ToolKit: A Versatile Software Suite for Analysis of High-Dimensional Free Energy Surfaces and Alchemical Free Energy Networks (2025) 65, 5273-5279 DOI: 10.1021/acs.jcim.5c00554 Read More View Full Article Download PDF |