Divalent Metal Ion Activation of a Guanine General Base in the Hammerhead Ribozyme: Insights from Molecular Simulations
Biochemistry vol. 56 p. 2985-2994 DOI: 10.1021/acs.biochem.6b01192
PMID/PMCID: PMC5832362 Published: 2017-06-12
Abstract
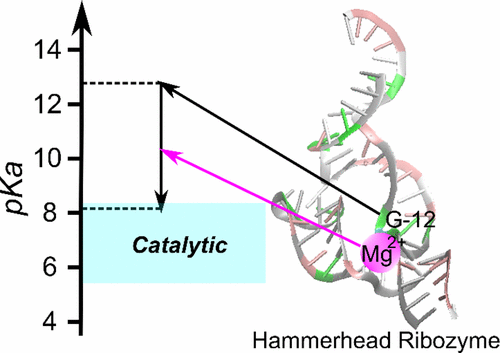
The hammerhead ribozyme is a well-studied nucleolytic ribozyme that catalyzes the self-cleavage of the RNA phosphodiester backbone. Despite experimental and theoretical efforts, key questions remain about details of the mechanism with regard to the activation of the nucleophile by the putative general base guanine (G12). Straightforward interpretation of the measured activity–pH data implies the pKa value of the N1 position in the G12 nucleobase is significantly shifted by the ribozyme environment. Recent crystallographic and biochemical work has identified pH-dependent divalent metal ion binding at the N7/O6 position of G12, leading to the hypothesis that this binding mode could induce a pKa shift of G12 toward neutrality. We present computational results that support this hypothesis and provide a model that unifies the interpretation of available structural and biochemical data and paints a detailed mechanistic picture of the general base step of the reaction. Experimentally testable predictions are made for mutational and rescue effects on G12, which will give further insights into the catalytic mechanism. These results contribute to our growing knowledge of the potential roles of divalent metal ions in RNA catalysis.