Toward Fast and Accurate Binding Affinity Prediction with pmemdGTI: An Efficient Implementation of GPU-Accelerated Thermodynamic Integration
Journal of Chemical Theory and Computation vol. 13 p. 3077-3084 DOI: 10.1021/acs.jctc.7b00102
PMID/PMCID: PMC5843186 Published: 2017-07-05
Abstract
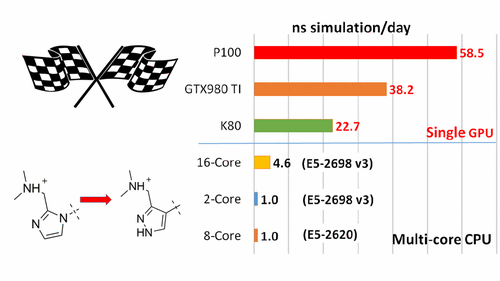
We report the implementation of the thermodynamic integration method on the pmemd module of the AMBER 16 package on GPUs (pmemdGTI). The pmemdGTI code typically delivers over 2 orders of magnitude of speed-up relative to a single CPU core for the calculation of ligand-protein binding affinities with no statistically significant numerical differences and thus provides a powerful new tool for drug discovery applications.