Multiscale Modeling
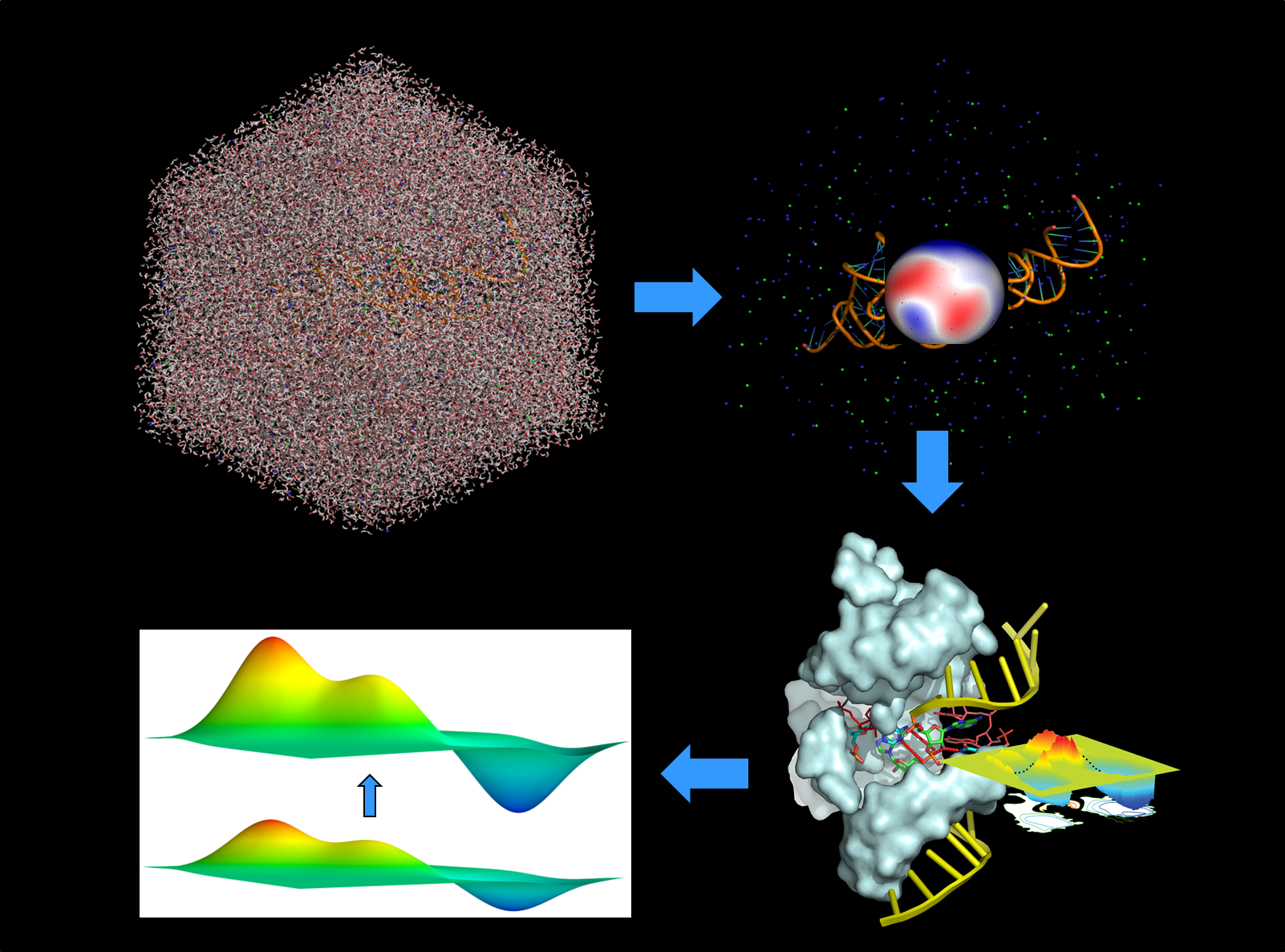
Multiscale modeling methods involve the integration of a hierarchy of theoretical methods that work synchronously or asynchronously to address complex chemical problems that span a large range of spatial and temporal scales. Example of a multiscale modeling strategy: Large-scale molecular dynamics simulations are performed to study the structure, dynamics and solvation of the extended enzyme system. This information is used to create a generalized solvation boundary that surrounds the active site and mimics the effect of the full solvated macromolecular system, but for a fraction of the computational cost. This reduced dimensional active site model is then used to conduct combined quantum mechanical/molecular mechanical (QM/MM) simulations with fast QM models, in conjunction with enhanced sampling and free energy methods, to construct the free energy landscape and pathway for the reaction. These landscapes are then refined to higher level QM methods using variational free energy expansion methods. The resulting free energy landscape and pathways for the reaction can then be used to aid in the interpretation of experiments, make experimentally testable predictions, and guide the design of new experiments or biotechnology. Elements of our multiscale modeling strategy include the development of new methods for:
- Combined quantum mechanical/molecular mechanical simulations
- Generalized electrostatics and solvation
- Free energy landscapes and pathways
Combined Quantum Mechanical/Molecular Mechanical Simulations
Combined quantum mechanical/molecular mechanical (QM/MM) methods are a practical approach to study complex chemical reactions in biological systems. In this approach, a relatively small localized region, such as the active site of an enzyme, is treated with a rigorous, and computationally demanding quantum mechanical model, whereas the rest of the system is treated using a much more computationally efficient molecular mechanics force field. This allows simulations to be performed of the chemical reaction itself, which ultimately may lead to a detailed understanding of catalytic mechanism. The York Lab develops and applies new combined QM/MM methods to study the catalytic mechanisms of protein and RNA enzymes.
Generalized Electrostatics and Solvation
Simulations of biological systems require consideration of not only biological macromolecules, but also their extended environment, which may be aqueous solution, a lipid membrane, a protein crystal, or the complex ionic atmosphere around and RNA molecule. "Generalized Electrostatics and Solvation" refer to this extended environment and the long-ranged forces that they exert. The York Lab develops methods to accurately and efficiently treat long-range electrostatic interactions and model generalized solvation so that simulations can be performed in these complex environments.
Free Energy Landscapes and Pathways
Free energy landscapes provide a survey map from which mechanistic pathways can be identified and quantified for chemical processes, conformational or binding events. The York Lab develops new methods to improve the sampling and construction of free energy surfaces, as well as methods to systematically correct these surfaces to more accurate levels at reduced computational cost.